DNA Sequencing methods and Strategies

Introduction:
DNA sequencing methods and strategies have become increasingly important in understanding and treating genetic diseases, of which there are estimated to be nearly 6000. These methods are also crucial for the production of beneficial compounds, such as anticancer drugs and hormones, as well as in fields such as genetic engineering, virology, metagenomics, and more. The advancement of DNA sequencing techniques continues to pave the way for more effective genetic engineering and therapies. DNA sequencing technique was able to sequence the entire human genome which helped in revealing many secrets lying in genes.
DNA sequencing:
“ DNA sequencing is the method of determining the order of nucleotide sequence of the DNA to gather up all the knowledge of an organism’s genetic makeup.”
This includes multiple complex techniques to determine the four bases- Adenine, Thymine, Cytosine, and Guanine. Where A pairs with T via two hydrogen bonds and G pairs with C with three hydrogen bonds encodes the entire genetic knowledge and determines the morphological characteristics of organisms. DNA sequencing has played a significant role in the Human genome project.
The strategy of DNA sequencing involves simple laboratory techniques to complex computational methods.
DNA Sequencing methods:
Chemical Degradation Method Of DNA sequencing
Sanger sequencing Method Of DNA sequencing
Pyrosequencing Method Of DNA sequencing
Whole Genome Shot Gun Sequencing Method Of DNA sequencing
Next-Gen Sequencing Method Of DNA sequencing
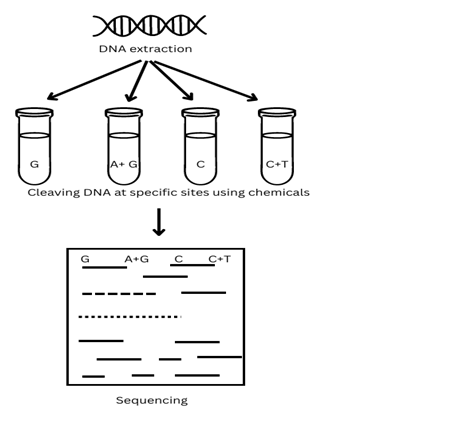
Chemical Degradation Method of Genome Sequencing: Given by Allan Maxam and Walter Gilbert. This method involves the chemical degradation of the sugar-phosphate backbone of a radio-labelled DNA molecule at specific sites.
The chemical modifications of base pairs or set base pairs followed by their cleavage. The visualization and sequencing of several base pairs or fragments produced are done via SDS-PAGE gel electrophoresis.
Procedure:
- Chemical degradation of Purified DNA fragments.
- Labelling of Single DNA strand with Phosphate32 by treating with alkaline phosphatase enzyme at 5’ end phosphate.
- Addition of P32 labelled ATP into the reaction mixture in presence of Phosphate kinase, which catalyzes the attachment of P32 at 5’end.
- The labelled DNA is then modified by treating it with different chemicals. The chemicals used are mainly dimethyl sulphate, hydrazine, and NaCl.
- The modified DNA is then incubated with piperidine to facilitate the cleavage of the sugar-phosphate backbone of DNA next to where it is modified.
- Visualization of modified fragments via SDS-PAGE.
Limitations:
- It requires the use of hazardous chemicals which might also destroy the native structure of DNA.
- Relatively complex and laborious technique.
- Slow and only limited to the length of 100bp in one reaction.
- Not suitable over the 500bp range.
- Not suitable for organisms with complex and large genomes like the human genome.
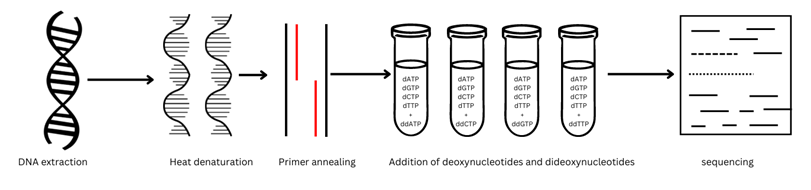
Sanger sequencing: This method is known as the Dideoxy or chain-termination method of DNA sequencing. This method was invented by Frederick Sanger in 1977 who was awarded the noble prize in 1980.
Sanger Sequencing method employs the addition of Dideoxy nucleotides (ddNTPs) in DNA sequence in vitro due to which the DNA replication terminates because of the inability of DNA polymerase to use dideoxy nucleotides for DNA synthesis, the reaction terminates when DNA polymerase encounters DDNTP’s which results in the production of DNA fragments which are then sequenced and visualized via Gel electrophoresis.
The ddNTPs are always added in 100-fold lower concentration than Deoxynucleotides. The deoxynucleotide added are labelled with P32 at their 5’end.
Method:
- The deoxynucleotides employed lack the 3’OH group at their end, so there is no phosphodiester bond formation and the reaction ceases.
- The DNA replication proceeded in four separate reactions with all four types of standard deoxynucleotides (dATP, dGTP, dTTP, dCTP), DNA primers and DNA polymerase.
- When the reaction is initiated add only one of four ddNTPs.
- All four ddNTPs (ddATP, ddGTP, ddTTP, ddCTP) are added sequentially into the reaction mixture.
- After completing the rounds of DNA replication, the DNA fragments are heat denatured and are separated according to their size via Gel electrophoresis.
- The DNA bands produced are visualized under a UV illuminator or by autoradiography.
- The position of the DNA bands obtained from the pattern is then used to read the sequence.
Advantages of the Sanger Sequencing Method:
- With the help of specifically designed oligonucleotides, DNA synthesis can be initiated at any point of choice.
- Radioactive labelling of DNA strands is not required.
- No frequent need for of denaturation DNA for single strands.
Improved Sanger Sequencing Method: The original sanger sequencing technique is modified in ways that are,
- Introduction of alkali denaturation of Plasmid DNA to eliminate heat denaturation process for the production of ssDNA.
- Previously the DNA polymerase I from E. coli was used but it comes with low specificity and processivity, So, the modification is done by introducing the DNA polymerase from bacteriophage T7 which is comparably more processive and sensitive.
- The use of enzyme which lacks the 5’-3’ exonuclease activity like Taq DNA polymerase for the production of more readable fragments and the use of Taq DNA polymerase is often combined with PCR amplification of DNA fragments.
- Labelling ddNTPs with fluorescent dye to avoid radioactive labels and for better visualization via laser-based detection.
- The modified method utilizes fluorescent labelling of the chain terminator ddNTPs, which permits sequencing in a single reaction, rather than four reactions as in the labelled primer method. In this sequencing method, each of the four DDNTP’s chain terminators is labelled with fluorescent dyes, each of which emits light at different wavelengths.
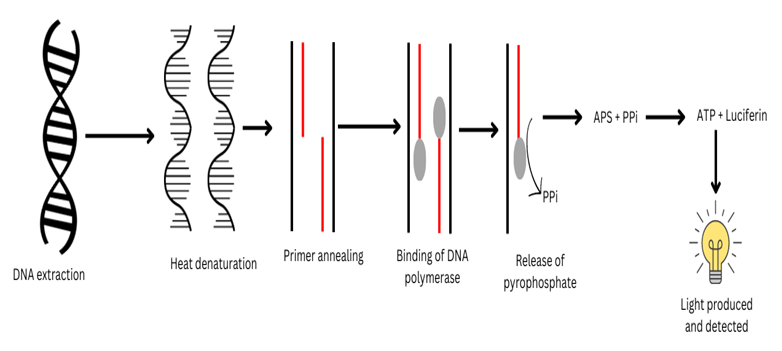
Pyrosequencing Method Of DNA sequencing: This method was invented by Pal Nyren in 1987 an advancement over Sanger Sequencing. He described that DNA sequencing can be done by determining the rate of DNA replication by measuring the pyrophosphate production, which can be detected by light.
Based on ‘Sequencing by Synthesis’ i.e., determining the rate of DNA molecule synthesis in real-time.
The method is based on the release of pyrophosphate when the new nucleotide is added to the DNA template and light is produced. By determining the release of pyrophosphate, we can determine the rate of replication and what nucleotide is added will help in determining the sequence of the DNA sample.
Method:
- It is based on the chain of enzymatic reactions that occur during DNA replication when the inorganic pyrophosphate is released due to nucleotide incorporation by DNA polymerase during primer-based DNA synthesis.
- The APS is supplemented into a reaction mixture (adenosine-5’-phosphosulfate) in the presence of DNA polymerase, When the reaction proceeds the pyrophosphate is released and gets converted to adenosine triphosphate (ATP) by the enzyme ATP sulfurylase.
- The ATP produced by the reaction is used as an energy source for the luciferin oxidation reaction catalyzed by the luciferase enzyme that generates light.
- The light is detected by a photodiode, photomultiplier tube, or a charge-coupled device (CCD) camera.
- This is followed by degradation of remaining ATP or any other dNTP by Apyrase enzyme treatment to allow the next round of sequencing reactions to be repeated to read the next base.
- Use of Klenow fragment of Escherichia coli DNA Pol I, ATP sulfurylase obtained from Saccharomyces cerevisiae, and the luciferase obtained from the American firefly Photinus pyralis is used in this method of DNA sequencing
Improved and Modified Version of Pyrosequencing: The improvement in the Pyrosequencing method is done and the Modified version is extensively used for Genome sequencing. DNA sequencing was enhanced by the use of Roche’s/454 titanium platform. The DNA amplified beads are loaded onto individual Picotiter Plate (PTP) wells, and additional beads, coupled with sulfurylase and luciferase, are added. The fibre-optic slide is mounted in a flow chamber, Sequencing reagents are delivered on the bead-packed wells. Fibre optic device is attached to High resolution-charged couple device camera from downward, which detects the release of light on the production of pyrophosphate during the pyrosequencing reaction. The light generated by the enzymatic reactions is recorded as a series of peaks called a flow gram and sequencing DNA.
Whole Genome Shotgun Sequencing: Whole genome shotgun sequencing is also an in vitro technique to determine the whole genome and whole exome of an organism. The DNA double helix molecule of an organism is broken down into fragments and is computationally assembled to determine the genetic knowledge of an organism and form library. Multiple reads are obtained when sequencing numerous small fragments of the target DNA molecule.
This technique was first described in 1979 and implied on the Cauliflower mosaic virus.
Steps involved in Whole Genome Shotgun Sequencing:
- The large genome of an organism is broken down into small fragments using ultrasonic waves. The DNA fragments are then purified.
- The purified small DNA fragments are then ligated with vector plasmid.
- The recombinant fragments are transformed into cells and cloned.
- The 1000’s DNA fragments are produced and sequenced using an automated computerized sequencer using Dye labelled primers.
- Using Computer software, the long stretch of DNA fragments are assembled using the shorter fragments by comparing nucleotide overlap sequences.
- The longer stretch is formed using the shorter fragments by filling the gaps and contig sequence. The Longer DNA stretch is produced are then proofread to resolve any ambiguity.
Advantages of Whole Genome Shotgun Sequencing:
- Increased speed and specificity.
- Fewer chances of errors.
- Improved accuracy of detection
- Increased length
- Can sequence longer stretches of DNA at one time.
- Improve sequence efficiency.
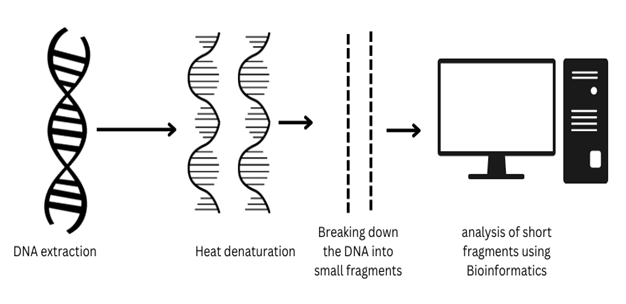
Next Generation Sequencing: Next Generation Sequencing is a massively parallel, advanced and deep technique of genome sequencing and RNA sequencing. Based on multiple methods of nucleotide sequencing. A rapidly advanced technique which can sequence a whole human genome within a single day. Illumina sequencing is one of the modified Next Generation sequencing techniques.
Bioinformatics plays a crucially important role in the sequencing of genomes in Next Generation Sequencing
Elaboration of Next Generation Sequencing is beyond the scope to date.
Four major steps in Next Generation Sequencing:
1. Isolation of Nucleic acid
2. Genome library preparation
3. Clonal amplification
4. Sequencing
5. Data analysis
Applications of Next-Generation Sequencing:
- NGS can detect a wide range of mutations than any other sequencing technique
- Genomes can be interrogated without bias
- The increased sensitivity of NGS allows the detection of mosaic mutations
- Microbiology and oncology
- Determine DNA protein interaction
Conclusion: The field of DNA sequencing is growing readily and day by day developing new and rapid techniques for sequencing the entire genome of organisms within the shortest period. With the upcoming era of new and advanced bioinformatics and metaverse also plays an important role in next genome sequencing. Next Generation Sequencing and other upcoming DNA sequencing techniques are much more complex but rapidly advance over previously described conventional techniques like Sanger sequencing, and pyrosequencing. A major future aspect required in this field of technology is the requirement of software or system which can read longer sequences in as short a period as possible. This is going to be a robust platform with multiple advantages in various fields like diagnostics, human genomics, molecular biology, genetics, industrial microbiology, metagenomics, gene therapy, evolutionary biology, recombinant DNA technology and treatment.
Important Questions:
Which was the first organism whose whole genome was sequenced and by which technique?
Bacteriophage phiX174 was the first organism whose genome was sequenced using the PacBio HiFi DNA sequencing method, it was chosen because it has a relatively small genome of few base pairs (48,502) by Sanger.
What are the Pros and Cons of Next Generation Sequencing?
Cons:
1)High speed and accuracy.
2)No chemical degradation of nucleotides.
3)Applicable for all organisms
Pros:
1)The main disadvantage of NGS is putting it in place requires a huge infrastructure, such as computer capacity and storage, and also required trained professionals for application.
2)Expensive
Which was the first prokaryotic organism sequenced and by which technique?
Haemophilus influenzae was the first prokaryotic organism whose DNA was sequenced in 1995 using the whole genome shotgun strategy.
What is a contig sequence?
A contig sequence is a set of overlapping DNA sequences in sequencing projects, which represents a consensus region of DNA.

![[Download] Jawetz Melnick and Adelberg’s Medical Microbiology PDF Book Download 9 [Download] Jawetz Melnick and Adelberg’s Medical Microbiology PDF Book](https://biologywala.com/wp-content/uploads/2022/08/Jawetz-Melnick-_-Adelbergs-Medical-Microbiology-PDF-Book-_-BIOLOGYWALA.COM_-392x245.webp)