Chromosome: level of DNA packaging

Chromosomes are thread-like, deeply staining structures found inside the nucleus of every eukaryotic cell. They represent the highest level of DNA packaging, ensuring that an extremely long DNA molecule fits within a microscopic nucleus.
The term “chromosome” (Greek: chroma = color, soma = body) was coined by W. Waldeyer (1888) because these structures take up certain stains vividly during cell division.
Each chromosome carries genes, the hereditary units responsible for the transmission of traits and control of cellular function.
2. Chemical Composition of Chromosomes
Chromosomes are made up of a nucleoprotein complex — primarily:
| Component | Percentage | Function |
| DNA | 30–40% | Genetic material; carries hereditary information |
| Histone proteins | 50–60% | Structural proteins responsible for DNA coiling |
| Non-histone proteins | 10–15% | Enzymatic and regulatory functions |
| RNA | 1–10% | Transcriptional activity (mRNA, rRNA, etc.) |
Histones are basic proteins (rich in lysine and arginine), whereas non-histone proteins are acidic, providing structural support and playing roles in replication and transcription.
3. Structure of Chromosome
Each metaphase chromosome is highly condensed and visible under a light microscope. The major parts are:
(a) Chromatid
Each duplicated chromosome consists of two identical strands called sister chromatids, joined at the centromere. Each chromatid has a complete DNA double helix.
(b) Centromere (Primary Constriction)
A constricted region that joins the sister chromatids. It is the site where spindle fibres attach during cell division via kinetochores.
Depending on the position of the centromere, chromosomes are classified into:
| Type | Centromere position | Shape |
| Metacentric | Middle | V-shaped |
| Submetacentric | Slightly off-center | L-shaped |
| Acrocentric | Near one end | J-shaped |
| Telocentric | At the terminal end | i-shaped |
(c) Secondary Constriction and Satellite
Some chromosomes show a secondary constriction apart from the centromere.
- Secondary constriction I: Forms nucleolar organizer regions (NORs) — responsible for rRNA synthesis.
- Secondary constriction II: A small fragment beyond it forms a satellite, giving rise to sat-chromosomes.
(d) Telomeres
Each chromatid ends with a telomere, a repetitive DNA sequence (TTAGGG in humans) that protects chromosome ends from degradation and prevents fusion with other chromosomes.
4. Molecular Organization – DNA Packaging in Chromosomes
The packaging of DNA into chromosomes occurs in a hierarchical, multilevel manner, ensuring condensation without tangling and maintaining accessibility for gene expression.
5. Levels of DNA Packaging
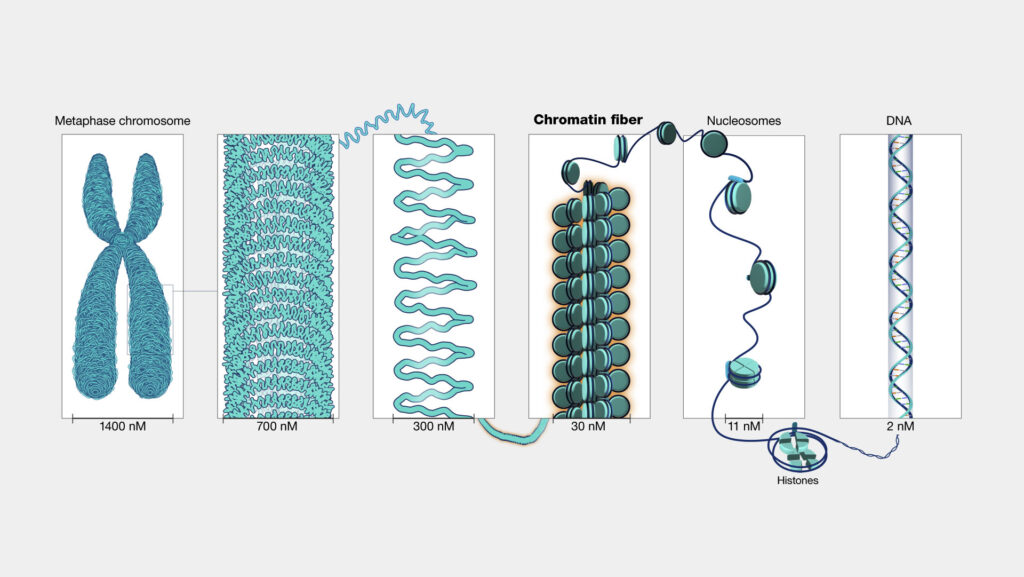
(a) Level 1 – Nucleosome (10 nm Fibre)
- The basic structural unit of chromatin discovered by Roger Kornberg (1974).
- Each nucleosome contains:
- Histone octamer (2 × H2A, 2 × H2B, 2 × H3, 2 × H4).
- 146 base pairs (bp) of DNA wrapped around the octamer in 1.65 turns.
- Linker DNA (~20–80 bp) connecting two adjacent nucleosomes.
- H1 histone acts as a clamp at the entry and exit point of DNA.
This structure appears as “beads on a string” under an electron microscope.
Each bead (nucleosome) ≈ 11 nm in diameter.
(b) Level 2 – Solenoid or 30 nm Fibre
- Nucleosomes coil further into a 30 nm thick fibre, known as the solenoid model (proposed by Finch & Klug, 1976).
- In this structure, 6 nucleosomes form a single turn of the solenoid.
- The histone H1 stabilizes this coiling.
- Represents the chromatin fibre during the interphase nucleus.
(c) Level 3 – Loop Domains (300 nm Fibre)
- The 30 nm solenoid fibre forms looped domains (40–100 kb of DNA) attached to a non-histone scaffold protein such as topoisomerase II and SMC proteins (Structural Maintenance of Chromosomes).
- These loops facilitate gene regulation, as active genes may be located at loop boundaries.
(d) Level 4 – Chromatid Fibre (700 nm)
- During prophase, the 300 nm fibre is further coiled to form a 700 nm chromatid.
- The loops coil and fold extensively, condensing DNA up to 10,000-fold from its extended length.
(e) Level 5 – Metaphase Chromosome (~1400 nm)
- The most compact form of DNA is seen during metaphase, when two sister chromatids (each 700 nm thick) are held together at the centromere.
- The DNA is compacted approximately 50,000 times compared to its extended length.
6. DNA Packaging Ratio
In humans:
- Total DNA length per cell ≈ 2 meters (2 × 10⁹ nm).
- Nuclear diameter ≈ 10 µm.
- Therefore, the packaging ratio (DNA length : chromosomal length) ≈ 1 : 50,000.
This high level of compaction is achieved without knotting, due to supercoiling and histone-mediated organization.
7. Supercoiling of DNA
Supercoiling refers to the overwinding or underwinding of DNA around its axis, allowing efficient packaging.
- Negative supercoiling (common in eukaryotes) facilitates unwinding during transcription and replication.
- Enzymes called topoisomerases (DNA gyrase) introduce and relax these supercoils.
8. Role of Histone and Non-Histone Proteins
| Protein Type | Function |
| Histones | Structural organization, forming nucleosomes |
| H1 (Linker histone) | Locks DNA in place and stabilizes nucleosomes |
| Non-histone proteins | Scaffold formation, gene regulation, replication, repair enzymes |
Histone modifications such as acetylation, methylation, phosphorylation, and ubiquitination alter chromatin compaction and are key in epigenetic regulation.
9. Euchromatin and Heterochromatin in Packaging
| Feature | Euchromatin | Heterochromatin |
| Compaction | Loosely packed | Densely packed |
| Transcriptional activity | Active | Inactive |
| Replication timing | Early S phase | Late S phase |
| Example | Gene-rich regions | Centromeres, telomeres |
10. Functional Significance of DNA Packaging
- Space efficiency – Long DNA fits into a microscopic nucleus.
- Protection – DNA is shielded from nuclease enzymes and damage.
- Regulation – Chromatin remodeling allows control of gene expression.
- Segregation – Highly condensed chromosomes ensure accurate distribution during cell division.
- Repair and replication – Proper chromatin organization aids enzymatic access during replication and DNA repair.
11. Summary Table: Levels of Chromatin Packaging
| Level | Structure | Diameter (nm) | DNA Length per Turn (bp) | Fold Compaction |
| 1 | DNA double helix | 2 | 10 bp per turn | 1× |
| 2 | Nucleosome | 11 | 146 bp per bead | ~7× |
| 3 | Solenoid fibre | 30 | 6 nucleosomes per turn | ~40× |
| 4 | Looped domains | 300 | 40–100 kb per loop | ~1000× |
| 5 | Chromatid | 700 | Highly condensed | ~10,000× |
| 6 | Metaphase chromosome | 1400 | Sister chromatids joined | ~50,000× |
The chromosome structure and DNA packaging represent a remarkable biological design balancing maximum condensation with regulated accessibility.
From the simple double helix to the complex metaphase chromosome, the hierarchy ensures efficient storage, protection, and function of the genetic material essential for life.
![[DOWNLOAD] Molecular and Cell Biology for Dummies PDF Book 3 [DOWNLOAD] Molecular and Cell Biology for Dummies PDF Book](https://biologywala.com/wp-content/uploads/2021/10/DOWNLOAD-Molecular-and-Cell-Biology-for-Dummies-PDF-Book-1-500x245.jpg)

