CSIR NET Life Science Dec 2024 Answer Key (Parts C) – Complete Explanations! Part 2

CSIR NET Life Science Dec 2024 Answer Key (Parts C) – Complete Explanations! Part 2
Q.1 The helicase activity of E. coli DnaB was investigated using the following two substrates (I and II) under various conditions, followed by gel electrophoresis and autoradiography. The results of these experiments are depicted below:
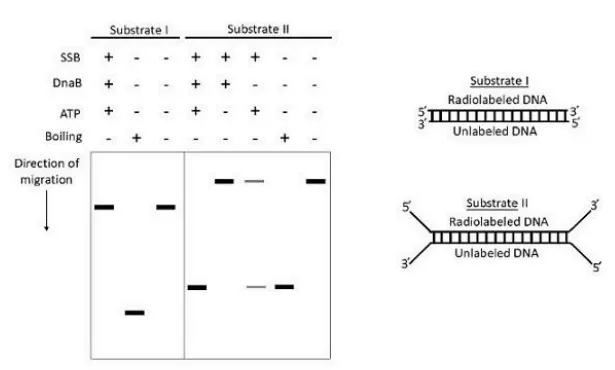
The following statements are made purely from the results shown above:
A. DnaB can unwind only a partially unwound DNA.
B. SSB inhibits the unwinding activity of DnaB.
C. DnaB unwinds DNA in the 5′ to 3′ direction.
D. DnaB requires ATP for DNA unwinding.
Which one of the following options represents the combination of all correct
statements?
- A, Band D
- A, C and D
- C and D only
- A and D only
Answer: 4
Explanation:
DnaB helicase needs ATP to function, and it acts at replication forks where DNA is already partially unwound. So, ATP is required (D), and it cannot start unwinding from completely double-stranded DNA (A). SSB (single-stranded binding protein) actually helps by stabilizing ssDNA, not inhibiting DnaB, so B is false. While DnaB does move 5’ to 3’, the gel data may not confirm this alone, so C is not included.
Q. 2 Two closely related sympatric ladybird beetle species in a rainforest have evolved to specialize on different insect prey. Which one of the following statements does NOT explain the speciation process in these beetle species?
- Populations exploited different diets in the rainforests.
- Over time, natural selection favored traits that allowed the consumption of distinct diets.
- Diverging populations developed differences in diet. However, it did not lead to reproductive isolation.
- Temporal differentiation in their foraging activity led to their distinct diets.
Answer: 3
Explanation:
For speciation to occur, reproductive isolation is essential. If two populations develop different diets but continue to interbreed, they have not become separate species. Therefore, this statement contradicts the process of speciation and is the correct answer.
Q. 3 Given below are a few statements regarding gene actions observed in plants.
A. In terms of pollination, self-pollinated species often exhibit additive gene action.
B. Non-additive gene action is less prevalent in cross-pollinated species.
C. Simply inherited (qualitative, oligogenic) traits predominantly exhibit non additive and epistatic gene action.
D. Genetic fixation of superior genes will be more difficult with dominance gene action.
Which one of the following options represents the combination of all correct statements?
- A, Band C
- A, C and D
- B, C and D
- A and B only
Answer: 2
Explanation:
Additive gene effects are common in self-pollinated plants due to limited genetic variation. Traits governed by few genes (qualitative traits) often show complex interactions like epistasis. Dominance gene action makes breeding harder because recessive alleles can be hidden, making it difficult to fix good traits. However, statement B is false — non-additive effects are actually more common in cross-pollinated species.
Q. 4 The table given below provides a comprehensive list of selected plant diseases (Column X) and possible causal pathogens (Column Y).
| Column X | Column Y |
|---|---|
| Plant Disease | Pathogen |
| A. Apple scab | I. Claviceps purpurea |
| B. Ergot of cereals | II. Erwinia amylovora |
| C. Fire blight | III. Meloidogyne spp. |
| D. Root-Knot | IV. Venturia inaequalis |
Which one of the following options represents all correct matches between Column X and Column Y?
- A-IV B-II C-III D-I
- A-II B-I C-IV D-III
- A-I B-IV C-III D-II
- A-IV B-I C-II D-III
Answer: 4
Explanation :
A. Apple scab — IV. Venturia inaequalis
B. Ergot of cereals — I. Claviceps purpurea
C. Fire blight — II. Erwinia amylovora
D. Root-Knot — III. Meloidogyne spp.
These are well-established disease-pathogen pairs. Apple scab is caused by a fungus, ergot is a fungal disease of cereals, fire blight is bacterial, and root-knot is caused by nematodes.
Q. 5 A researcher obtained a loss-of-function mutant of plasmodesmata protein synaptotagmin (SYTA) in Arabidopsis and infected the plants with cabbage leaf curl virus (CaLCuV). The following statements represent possible outcomes of the above experiment.
A. The CaLCuV infection will be slower in the mutant plants.
B. The mutant plants will be completely resistant to the viral infection.
C. The endocytic recycling pathway in the infected cells will be compromised in the mutant plants.
D. The disease symptoms will be more severe in the mutant plants.
Which one of the following options represents all correct statement(s)?
- A and C
- Band C
- B only
- D
Answer: 1
Explanation:
SYTA helps viruses move through plasmodesmata. Without it, the virus spreads more slowly (A). It’s not total resistance, just reduced efficiency. Also, SYTA is involved in membrane trafficking, so endocytic recycling is disrupted (C). B and D are not supported.
CSIR NET Life Science Dec 2024 Answer Key (Parts C) – Complete Explanations! Part 2
Q. 6 If a 0.1 M solution of glucose 1-phosphate is incubated with a catalytic amount of phosphoglucomutase, the glucose 1-phosphate is transformed to glucose 6- phosphate. At equilibrium, the concentrations of the reaction components are:
Glucose 1-phosphate ⇌ Glucose 6-phosphate
(4.5 X 10-3 M) (9.6 X 10-2 M)
What would be the calculated values for K’eq and ΔG°’ for this reaction at 25°C?
- (0.0045 to 0.096) and (-0.7 to -0.8) kJ/mol
- (21 to 22) and (-7.5 to -7.7) kJ/mol
- (0.1 to 0.2) and (-17.6 to -17.7) kJ/mol
- (21 to 22) and (-27.7 to -27.8) kJ/mol
Answer: 2
Explanation :
Step 1: Calculate the equilibrium constant (Keq)
Keq = [G6P] / [G1P] = 0.096 / 0.0045 ≈ 21.3
This means the reaction favors the formation of glucose 6-phosphate.
Step 2: Calculate ΔG°’
Use the formula: ΔG°' = –RT ln(Keq)
Where:
R = 8.314 J/mol·K
T = 298 K (room temperature)
ln(21.3) ≈ 3.06
Plug in the values:
ΔG°’ = –(8.314)(298)(3.06) ÷ 1000 ≈ –7.6 kJ/mol
Final Answer:
Keq ≈ 21
ΔG°’ ≈ –7.6 kJ/mol
The negative ΔG°’ means the reaction goes forward and releases energy.
Q. 7 A purified 150 kDa protein species from gel filtration column was run on a 2- dimensional SDS-PAGE as shown below:
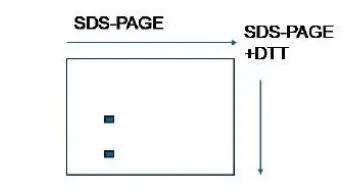
What is the likely form of the 150 kDa protein species from this observation?
- There are at least two proteins that are linked through non-covalent interactions.
- There are at least two proteins in the complex that are linked through covalent bonds.
- There are two proteins in the mixture without forming a complex.
- There is only one protein that has a disulfide bond .
Answer: 2 or 4
Explanation:
Gel filtration gives the native size (whole complex), but SDS-PAGE breaks subunits apart. If multiple spots appear, it means the protein is made of more than one subunit. If they stay together even under SDS, they are likely covalently linked, such as by disulfide bonds.
Q.8 Given below are different types of genetic manipulations of E. coli trp operon (Column X) and their consequences on its transcription (Column Y) under high tryptophan concentration.
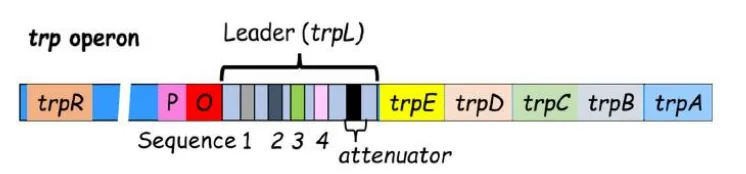
| Column X | Column Y |
|---|---|
| A. Inserting bases between the leader peptide gene and sequence 2 | i. Complete attenuation |
| B. Inserting bases between sequences 2 and 3 | ii. Less attenuation |
| C. Deleting sequence 4 | iii. More attenuation |
| D. Elimination of ribosome-binding site for the leader peptide | iv. No attenuation |
Which one of the following options represents all correct matches between Column X and Column Y?
- A (i) B (ii) C (iii) D (iv)
- A (i) B (iii) C (ii) D (iv)
- A(iv) B (ii) C (iii) D (i)
- A (ii) B (iii) C (iv) D (i)
Answer: 4
Explanation:
A. Bases inserted between leader and seq 2 → ii. Less attenuation
B. Between seq 2 and 3 → iii. More attenuation
C. Delete seq 4 → iv. No attenuation
D. No RBS for leader peptide → i. Complete attenuation
Each structural change alters RNA secondary structure, affecting the formation of termination (attenuation) hairpins. Deletion of sequence 4 removes the terminator stem-loop, preventing transcription termination via attenuation. If the leader peptide cannot be translated—such as due to a missing ribosome binding site (RBS)—the ribosome stalls, allowing the terminator hairpin to form, and attenuation occurs by default. Insertions at different locations influence which regions of the mRNA can base pair, thereby altering which hairpin structures form and whether transcription is terminated or continues.
Q. 9 Lethally irradiated mice were split into 4 groups and experiments were conducted as described below:
Group 1 was not given any cells.
Group 2 was given thymus-derived cells from a syngeneic donor, and two months later, immunized with a polysaccharide antigen.
Group 3 was given bone marrow cells from a syngeneic donor, and two months later, immunized with a polysaccharide antigen.
Group 4 was given bone marrow cells from a syngeneic donor, and two months later, immunized with a T-dependent antigen.
Four possible outcomes, listed below, were suggested.
A. Group 1 mice are unlikely to survive.
B. Group 2 mice are likely to produce antibodies in response to polysaccharide antigens.
C. Group 3 mice are likely to produce antibodies in response to polysaccharide antigens.
D. Group 4 mice are likely to produce antibodies in response to T-dependent antigens.
Which one of the following options represents the combination of all correct
statements?
- A and B only
- A, B and D
- A and C only
- A, C and D
Answer: 4
Explanation:
Mice with no cells (Group 1) die due to loss of immunity. Group 2, with only thymus cells, can’t make B cells, so they don’t respond to antigens. Group 3 gets bone marrow and can respond to polysaccharide antigens (B cell-dependent). Group 4 can respond to T-dependent antigens because both B and T cells are restored via bone marrow.
Q.10 Which statement about studies on succession is INCORRECT?
- Succession studies can reveal the effects of non-native species on the ecological structures and functions of a community.
- Studies of succession can indicate threshold conditions for the ‘invasion window’.
- Non-native invasion can divert succession by out-competing existing species.
- Succession studies show that coevolved native species always outcompete invasive species.
Answer: 4
Explanation:
This is incorrect because invasive species can sometimes dominate ecosystems, even over coevolved natives. Succession studies actually help understand how invasions can change community dynamics, but they don’t guarantee that natives will always win.
Also read :
CSIR NET Life Science Dec 2024 Answer Key (Parts C) – Complete Explanations! Part 3
CSIR NET Life Science Dec 2024 Answer Key (Parts C) – Complete Explanations! Part 4
CSIR NET Life Science Dec 2024 Answer Key (Parts C) – Complete Explanations! Part 5
CSIR NET Life Science Dec 2024 Answer Key (Parts C) – Complete Explanations! Part 6
CSIR NET Life Science Dec 2024 Answer Key (Parts C) – Complete Explanations! Part 1
CSIR NET Life Science Previous Years Question Papers and answer keys
Join SACHIN’S BIOLOGY on Instagram or Facebook to receive timely updates and important notes about exams directly on your mobile device. Connect with Mr. Sachin Chavan, the founder of Sachin’s Biology and author of biologywala.com, who holds an M.Sc., NET JRF (AIR 21), and GATE qualifications. With SACHIN’S BIOLOGY, you can have a direct conversation with a knowledgeable and experienced.

![[PDF] Plant Tissue Culture: Definition, Method, examples, advantages| Biology PDF Notes 6 [PDF] Plant Tissue Culture: Definition, Method, examples, advantages| Biology PDF Notes](https://biologywala.com/wp-content/uploads/2023/06/plant-tissue-culture-process-1-520x245.webp)
![[DOWNLOAD] Complete notes on Chromatin Organisation PDF](https://biologywala.com/wp-content/themes/hueman/assets/front/img/thumb-medium-empty.png)