CSIR NET Life Science Dec 2024 Answer Key (Parts C) – Complete Explanations! Part 6

CSIR NET Life Science Dec 2024 Answer Key (Parts C) – Complete Explanations! Part 6
Q. 1 Protein X can be extracted from disrupted erythrocyte plasma membranes with high salt concentrations. Treatment of intact erythrocytes with protease followed by extraction led to intact protein X. Treatment of disrupted erythrocyte plasma membranes with protease followed by extraction led to fragmented protein X. The following interpretations were made:
A. Protein X is a peripheral membrane protein
B. Protein X is an integral membrane protein
C. Protein Xis on the extracellular matrix face of the plasma membrane
D. Protein X is on the cytosolic face of the plasma membrane
Which one of the following options best represents the combination of all correct
interpretations?
- A and C
- A and D
- B and C
- B and D
Answer – 2
Explanation:
- Protein X can be extracted with high salt, which suggests it is a peripheral membrane protein (not tightly embedded like integral proteins).
- Protease treatment of intact erythrocytes does not digest Protein X, implying it’s on the cytosolic face (protease cannot access the inner face in intact cells).
- Protease treatment of disrupted membranes leads to fragmentation, confirming its exposure once the membrane is compromised.
- Thus, A (peripheral) and D (cytosolic face) are correct.
Q. 2 The muscle spindles are the stretch receptors that initiate stretch reflex in skeletal muscles. The following statements are proposed to describe the structural and functional characteristics of the different components of a muscle spindle.
A. The specialized intrafusal fibers in muscle spindles have non-contractile polar ends and a contractile centre.
B. The intrafusal fibers do not contribute to the overall contractile force of the muscle.
C. The primary sensory ending in a muscle spindle is formed by group la afferent fibers.
D. The axons of a-motor neurons having a diameter of 12-20 µm innervate the muscle spindles as the motor nerve.
Which one of the following options represents the combination of correct statements?
- A and B
- Band C
- C and D
- A and D
Answer – 2
Explanation:
- Statement A is incorrect: Intrafusal fibers have contractile polar ends and non-contractile center, not the other way around.
- B is correct: Intrafusal fibers help in proprioception but don’t generate contractile force.
- C is correct: Group Ia afferents form primary sensory endings.
- D is incorrect: α-motor neurons innervate extrafusal fibers, not the spindle. γ-motor neurons innervate intrafusal fibers.
Q. 3 Pollen tube growth in the transmission tract (TT) of the style and its attraction to the embryo sac are directed by chemical cues in plants, as discovered in Lily and Torenia. Which one of the following statements about the proteins/peptides secreted in the process is correct?
- TT epidermis secretes cysteine-rich defensin-like peptides, whereas synergids (SY) secrete cysteine-rich adhesins.
- TT epidermis secretes cysteine-rich adhesins, whereas SY secrete cysteine-rich defensin-like peptides.
- TT epidermis secretes proline- and hydroxyproline-rich glycoproteins whereas SY secrete cysteine-rich adhesin and chemocyanin.
- TT epidermis secretes arabinogalactan proteins and chemocyanin, whereas the central cell of the embryo sac secretes cysteine-rich LURE peptides.
Answer – 2
Explanation:
- The transmitting tract (TT) epidermis produces adhesive proteins (like arabinogalactan proteins, chemocyanin) that guide pollen tube growth.
- Synergid cells secrete LURE peptides, which are defensin-like cysteine-rich peptides that attract the pollen tube.
- So, the correct matching is: TT → adhesins; Synergids → defensin-like peptides.
Q. 4 A researcher simultaneously inhibited the activities of Triose-Phosphate Translocator (TPT) and Xylulose 5-Phosphate Translocator (XPT) in a plant and
made the following assumptions:
A. Triose phosphate will be accumulated more in the chloroplast.
B. Triose phosphate will be accumulated more in the cytosol.
C. Xylulose 5-phosphate will be accumulated more in the chloroplast.
D. Xylulose 5-phosphate will be accumulated more in the cytosol.
Which one of the following combinations of the above assumptions is correct?
- A and C
- A and D
- B and C
- B and D
Answer – 2
Explanation:
- TPT transports triose phosphate from chloroplast to cytosol.
- XPT transports xylulose-5-phosphate out of the chloroplast.
- If both are inhibited, these metabolites accumulate in the chloroplast, not cytosol.
- So, triose-P and xylulose-5-P both increase in the chloroplast, hence A and C.
Q. 5 Following statements are made regarding methyl erythritol phosphate (MEP) and mevalonate pathways for plant terpenoid biosynthesis.
A. The MEP pathway occurs in plastids whereas mevalonate pathway occurs in cytosol.
B. The mevalonate pathway supplies most of the C5 units of terpenoids for the biosynthesis of monoterpenes and diterpenes .
C. The MEP pathway supplies most of the Cs units of terpenoids for the biosynthesis of sesquiterpenes.
D. Certain genes of the plant MEP pathway are believed to have been acquired from the cyanobacterial symbiont.
Which one of the following options represents a combination of all correct statements?
- A, B and C
- B, C and D
- C and D only
- A and D
Answer – 4
Explanation:
- MEP pathway → plastids; Mevalonate pathway → cytosol (A is correct).
- Monoterpenes & diterpenes come mostly from MEP, not mevalonate (So, B is incorrect).
- Sesquiterpenes come mainly from mevalonate, not MEP (So, C is incorrect).
- D is correct: MEP pathway has genes likely acquired from cyanobacterial endosymbiont (due to plastid origin).
Q. 6 A student studying tree species diversity uses a large number of sampling quadrats (each of 1-hectare area) to cover >50% of the area of a 200-hectare tropical forest patch. Consider the statements in the options below:
A. Species numbers increase with sampling area following a power-law relationship with exponent >0 and <1.
B. The log of species numbers increases linearly with the log of the sampling area.
C. Species numbers increase with sampling area following a power-law relationship with exponent >1.
D. Species numbers increase linearly with the log of the sampling area.
Which combination of the statements above describes the expected pattern?
- A and B
- C only
- B and C
- A and D
Answer – 1
Explanation –
Correct Answer: A and B
The Species-Area Relationship (SAR) is a well-known ecological principle that states:
“As the area sampled increases, the number of species observed also increases.”
Mathematical Form –
The relationship usually follows a power-law:
S = c × Az
S= number of speciesA= area sampledc= constant (depends on environment)z= exponent (usually between 0.2 and 0.4)
Explanation of Correct Options –
A. Species numbers increase with area following a power-law with exponent >0 and <1
This is a direct description of the equation. Since 0 < z < 1, species richness increases sub-linearly with area.
B. The log of species numbers increases linearly with the log of sampling area
Taking the logarithm of the power-law equation:
log(S) = log(c) + z × log(A)
This gives a straight-line relationship between log(S) and log(A).
Why the Other Options are Incorrect –
C. Exponent >1 – Unrealistic; this would suggest species increase faster than area.
D. Species numbers increase linearly with log(area) – This would suggest a logarithmic or exponential trend, which doesn’t fit SAR.
Q. 7 The following statements describe a few basic features of the interferometric reflectance imaging sensor used as a biosensing platform:
A. This biosensing platform is capable of high-throughput multiplexing of protein-protein, protein-DNA and DNA-DNA interactions.
B. The sensing surface is prepared by robotic spotting of biological probes that are immobiliized on functionalised Si/Si02 substrate.
C. As biomass accumulates on the substrate surface, a change in the interferometric signature occurs and the change can be correlated to a quantifiable mass.
D. Using this technique, picometer changes in biomass may be detected.
Which one of the following options represents the combination of all correct statements?
- B, C and D
- C and D only
- A only
Answer – 1
Explanation:
The Interferometric Reflectance Imaging Sensor, or IRIS, is a special tool used to study how different biological molecules, like proteins and DNA, interact with each other. Let’s go through each statement.
Statement A:
“This biosensing platform is capable of high-throughput multiplexing of protein-protein, protein-DNA and DNA-DNA interactions.”
This is correct.
IRIS can detect many types of molecule interactions all at once. This is called multiplexing. It helps scientists test many samples quickly and efficiently.
Statement B:
“The sensing surface is prepared by robotic spotting of biological probes that are immobilized on functionalised silicon or silicon dioxide substrate.”
This is correct.
The surface of the IRIS sensor is made of a special material, like silicon. Tiny spots of biological molecules (called probes) are placed on the surface using robots. This makes sure the spots are placed accurately and consistently.
Statement C:
“As biomass accumulates on the substrate surface, a change in the interferometric signature occurs and the change can be correlated to a quantifiable mass.”
This is correct.
When other molecules stick or bind to the probes on the surface, they add more mass. IRIS detects this by measuring how light reflects off the surface. When the mass changes, the light reflection also changes. This allows scientists to calculate how much mass was added.
Statement D:
“Using this technique, picometer changes in biomass may be detected.”
This is incorrect.
IRIS is very sensitive and can detect very small changes in mass. However, it usually detects changes in the nanometer or smaller range, not at the picometer level. A picometer is much smaller, and IRIS is not typically accurate to that level.
Q.8 A researcher uses taxon weighting and complementarity as criteria to prioritise communities for biodiversity conservation. The diagram below shows the distributions of five taxa (A to E) among four regions (R1 -R4 ). Column W represents the weightage given to these five taxa based on their taxonomic uniqueness.
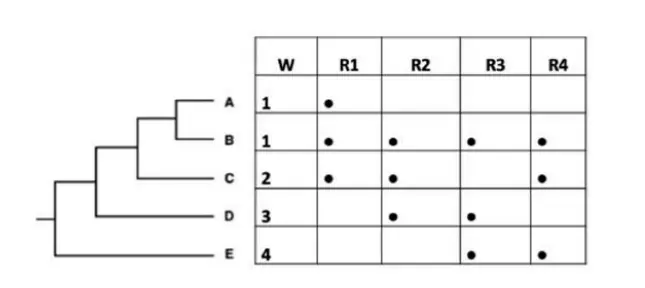
Select the option that lists the appropriate order of regions that should be prioritised (from highest to lowest) for conservation.
- R 1 > R3 > R4 > R2
- R2 > R1 > R3 > R4
- R3 > R 1 > R4 > R2
- R4 > R2 > R1 > R3
Answer – 3
Explanation:
To prioritize areas for conservation, two key criteria are used:
1. Taxon Weight (Uniqueness or Rarity):
Each species (taxon) is given a weight based on how unique or rare it is.
- A species found in only one region is given a high weight.
- A common species found in many regions is given a low weight.
Regions that contain more rare or unique species are considered more valuable.
2. Complementarity (Non-redundant Contribution):
This means how much new biodiversity a region adds compared to regions already selected.
- If a region has species that are not found in other regions, it adds more value.
- Regions with overlapping species (already protected elsewhere) are less prioritized.
How It Works in the Given Question:
- The researcher uses a matrix showing which species are found in which region, and a column showing species weight (rarity).
- By comparing this information, the region that adds the most rare and new species is ranked highest.
Why R3 > R1 > R4 > R2 is Correct:
- Region R3 has species with the highest uniqueness and least overlap with others.
- Region R1 adds valuable species after R3.
- R4 and R2 add fewer or more common species, so they are lower in priority.
Q. 9 The following statements were made regarding the roles of histone modifications in transcriptional regulation.
A. Acetylation of histones is generally associated with transcriptional repression by making the chromatin more compact.
B. Methylation of histones can either activate or repress transcription depending on the specific residue modified.
C. Phosphorylation of histones occurs in response to DNA damage and can influence gene expression.
D. Histone modifications influence the recruitment of RNA polymerase complex but not transcription factors .
Which one of the following options represents the combination of all correct statements?
- A and D only
- B and C only
- B, C, and D
- A, B, and D
Answer – 2
Explanation:
- A is incorrect: Acetylation activates transcription by loosening chromatin.
- B is correct: Methylation can either activate (e.g., H3K4me) or repress (e.g., H3K9me).
- C is correct: Phosphorylation is involved in DNA damage response and gene expression regulation.
- D is incorrect: Histone modifications can influence recruitment of both RNA polymerase and transcription factors.
Q. 10 The evolution of algal lineages is closely linked to endosymbiotic events. Which of the following statements best explains the origin and diversification of plastids in different algal groups?
- Primary plastids originated from a eukaryotic host cell engulfing a red algal ancestor, followed by secondary endosymbiosis leading to the diversification of green and brown algal lineages.
- Primary plastids evolved through the engulfment of a cyanobacterium by a eukaryotic host, while secondary and tertiary endosymbiosis involving red and green algae gave rise to plastids in diverse algal lineages such as diatoms and dinoflagellates.
- All algal lineages acquired plastids through multiple independent primary endosymbiosis events, with, cyanobacteria being engulfed by both red and
green algal ancestors. - Secondary endosymbiosis was responsible for the origin of primary plastids
in green algae, while tertiary endosymbiosis involving diatoms led to the
evolution of plastids in red algae.
Answer – 2
Explanation:
- Primary endosymbiosis: Eukaryote engulfed cyanobacterium → gave rise to red, green algae, and glaucophytes.
- Secondary endosymbiosis: Eukaryotes engulfed red/green algae → led to diatoms, euglenoids, dinoflagellates, etc.
- Tertiary endosymbiosis: Some protists further engulfed algae with secondary plastids.
Also read :
CSIR NET Life Science Dec 2024 Answer Key (Parts C) – Complete Explanations! Part 7
CSIR NET Life Science Dec 2024 Answer Key (Parts C) – Complete Explanations! Part 5
CSIR NET Life Science Dec 2024 Answer Key (Parts C) – Complete Explanations! Part 3
CSIR NET Life Science Dec 2024 Answer Key (Parts C) – Complete Explanations! Part 2
CSIR NET Life Science Dec 2024 Answer Key (Parts C) – Complete Explanations! Part 1
CSIR NET Life Science Previous Years Question Papers and answer keys
Join SACHIN’S BIOLOGY on Instagram or Facebook to receive timely updates and important notes about exams directly on your mobile device. Connect with Mr. Sachin Chavan, the founder of Sachin’s Biology and author of biologywala.com, who holds an M.Sc., NET JRF (AIR 21), and GATE qualifications. With SACHIN’S BIOLOGY, you can have a direct conversation with a knowledgeable and experienced.