CSIR NET Life Science Dec 2024 Answer Key (Parts C) Part 3

CSIR NET Life Science Dec 2024 Answer Key (Parts C) – Complete Explanations! Part 3
The table below shows types of chemical mutagens and names of mutagens.
Which one of the following options shows the correct match between Column X and Column Y?
A: iv B: iii C: i D: ii
A: i B: iv C: iii D: ii
A: ii B: iii C: iv D: i
A: iii B: ii C: i D: iv

Answer: 1
Explanation:
Mutagens are classified based on the type of chemical alteration they cause in DNA. Here’s the correct match:
A (Alkylating Agents – iv): Add alkyl groups to bases, often leading to mispairing.
B (Intercalating Agents – iii): Insert between DNA bases, causing frameshift mutations.
C (Base Analogs – i): Mimic normal DNA bases, incorporated during replication.
D (Deaminating Agents – ii): Remove amino groups from DNA bases, causing point mutations.
At what range of substrate concentration will an enzyme with a Kcat of 30 s-1 and a Km of 0.005 M show one-quarter of its maximum rate?
1. 3. 0 x 1 0-3 M to 3. 1 x 1 Q-3 M
2. 0.65 x10-3 M to 0.75 x10-3 M
3. 1.65 X 10-3 M to 1. 75 X 10-3 M
4. 2.7 x1 o-3 M to 2.8 x10-3 M
Answer: 3
Explanation:
🧪 Enzyme Kinetics: Finding Substrate Concentration for One-Quarter Vmax
To find the substrate concentration [S] at which an enzyme reaches one-quarter (¼) of its maximum rate (Vmax), we use the Michaelis-Menten equation:v = (Vmax × [S]) / (Km + [S])
Given:
Kcat = 30 s⁻¹ (not needed directly)
Km = 0.005 M = 5 × 10⁻³ M
v = ¼ Vmax
Step-by-step Calculation:
Substitute into the Michaelis-Menten equation:(1/4) Vmax = (Vmax × [S]) / (Km + [S])
Cancel Vmax from both sides:1/4 = [S] / (Km + [S])
Multiply both sides by (Km + [S]):(1/4)(Km + [S]) = [S]
Distribute and solve:(Km/4) + ([S]/4) = [S]
Multiply all terms by 4:Km + [S] = 4[S]
Simplify:Km = 3[S] → [S] = Km / 3 = (5 × 10⁻³) / 3 = 1.67 × 10⁻³ M
✅ Correct Option:
Option 3: 1.65 × 10⁻³ M to 1.75 × 10⁻³ M
A cytoplasmic monomeric protein containing a single non-surface exposed cysteine residue precipitates upon mutation of the Cys to ‘Ile’. However, the mutation of Cys to ‘Ala’ leads to a soluble and functional protein equivalent to the native form. Which one of the following statements explains the above observations?
1 . Cys mutated to ‘Ile’ alters the net charge of the protein, while mutation to ‘Ala’ does not.
2. Cys mutated to ‘Ala’ alters the net charge of the protein, while mutation to ‘Ile’does not.
3. Cys mutated to ‘Ala’ causes steric hindrance in the core of the protein, while
mutation to ‘lie’ does not.
4. Cys mutated to ‘lie’ causes steric hindrance in the core of the protein, while
mutation to ‘Ala’ does not.
Answer: 4
Explanation:
Cysteine can form disulfide bonds or maintain tight structural packing.
Isoleucine is bulkier and hydrophobic, causing steric hindrance in the protein core, disrupting folding.
Alanine is small and non-polar, maintaining structural integrity.
Given below are a few statements about intracellular protein transport.
A. Proteins that are destined for the lysosome are tagged with a mannose-6-
phosphate (M6P) group in the Golgi apparatus, which is recognized by the
M6P receptor in the trans-Golgi network.
B. Signal recognition particle directly mediates the insertion of proteins into the mitochondrial membrane.
C. The KDEL receptor in the ER and Golgi apparatus works by retrieving soluble ER resident proteins that have accidentally moved to the Golgi.
D. Cargo proteins that need to be exported from the ER are packaged into COPII vesicles based on the presence of an ER export signal in their cytosolic tail.
E. Clathrin-coated vesicles are primarily involved in vesicle trafficking between the Golgi apparatus and the ER.
Choose the option that has all correct statements.
1. A, C and D
2. A, Band E
3. A and D only
4. C and E
Answer: 1
Explanation:
A: Lysosomal proteins are tagged with mannose-6-phosphate (M6P) in the Golgi.
C: KDEL receptor retrieves ER-resident proteins from Golgi.
D: COPII vesicles export proteins from ER using cytosolic export signals.
B is incorrect — SRP guides proteins to ER, not mitochondria.
E is incorrect — Clathrin primarily functions in endocytosis and Golgi to endosome, not Golgi to ER.
The following statements are made regarding the role of cadherins in cell junctions.
A. Cadherins are calcium-dependent adhesion molecules that mediate cell to
cell adhesion by forming homophilic interactions.
B. Cadherins function in tight junctions, sealing the space between adjacent
cells.
C. N-cadherins are primarily found in epithelial cells and mediate adhesion to
the basal lamina.
D. Cadherins are involved in the formation of focal adhesiions linking the cell
cytoskeleton to the ECM.
E. Cadherins interact with the actin cytoskeleton through associated proteins
like catenins.
Which one of the following options represents all correct statements?
1. B, C and E
2. A, Band D
3. A and D only
4. A and E only
Answer: 4
Explanation:
A: Cadherins mediate calcium-dependent homophilic cell adhesion.
E: Cadherins link to actin cytoskeleton via catenins.
B, C, D are incorrect:
Tight junctions (B) involve claudins, not cadherins. Tight junctions are primarily formed by claudins and occludins, which seal the space between adjacent cells.
N-cadherins (C) are common in neurons, not basal lamina adhesion. They mediate cell-to-cell adhesion, particularly in tissues like the nervous system, not adhesion to the basal lamina.
Focal adhesions (D) use integrins, not cadherins. Integrins link the cell cytoskeleton (actin filaments) to the extracellular matrix (ECM), whereas cadherins are involved in cell-to-cell adhesion.
Given below are components that facilitate transfer of molecules across
phospholipid bilayers (Column X) and the properties of these components (Column Y).
Choose the option that correctly matches the components with their properties.
1. A (IV) B (II) C (I) D (Ill)
2. A (II) B (Ill) C (IV) D (I)
3. A (Ill) B (IV) C (I) D (II)
4. A (Ill) B (IV) C (II) D (I)

Answer: 4
Explanation:
Match the component type with function:
A (Channel proteins – III): Provide a hydrophilic pore.
B (Carrier proteins – IV): Undergo conformational changes for transport.
C (Pumps – II): Use ATP to transport substances against gradient.
D (Simple diffusion – I): Passive movement along gradient.
The following table shows forest floor litter pool and aboveground litterfall data for three forest types.
Based on the provided information, which one of the following options accurately
identifies the various forest types?
1. A- Tropical forest, B- Temperate deciduous forest, C- Temperate coniferous
forest
2. A- Temperate coniferous forest, B- Temperate deciduous forest, C- Tropical
forest
3. A- Temperate coniferous forest, B- Tropical forest, C- Temperate deciduous
forest
4. A- Tropical forest, B- Temperate coniferous forest, C- Temperate deciduous
forest
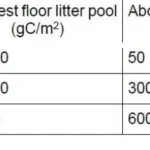
Answer: 2
Explanation:
Litterfall refers to the organic material that falls from the trees and other plants to the forest floor. This includes leaves, needles, fruits, seeds, and twigs. Litterfall is a measure of how much organic material is being added to the forest floor annually.
Litter pool is the accumulated layer of dead organic matter (the litter) that remains on the forest floor. It consists of all the decaying organic material that has not yet decomposed. The litter pool includes the material from previous years’ litterfall that has not fully decomposed.
Tropical forests: High litterfall, low litter accumulation due to rapid decomposition.
Temperate coniferous forests: High litter pool, slow decomposition (low temperature).
Temperate deciduous: Intermediate between tropical and coniferous.
Quantitative Trait Loci (QTLs) can be identified using two main approaches: biparental matings (BPM) and Genome-Wide Association Studies (GWAS). Below are some descriptors related to these strategies:
A. Large sample size
B. Small sample size
C. Population derived from controlled crosses
D. Random mating populations
E. Limited to two alleles per locus
F. Multiple alleles per locus
G. Linkage-based mapping
H. Linkage disequilibrium-based mapping
The table below presents four incomplete statements regarding BPM and GWAS. Each statement can be completed using the above descriptors.
Which one of the following options correctly completes all statements?
1. i
2. ii
3. iii
4. iv

Answer: 1
Explanation:
The table below shows different developmental processes and associated signaling molecules/pathways.
Which one of the following options represents the correct association of
developmental processes and signaling molecules?
1. A and B
2. B and D
3. A and C
4. C and D
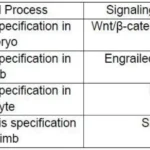
Answer: 1
A (Amphibian embryo D/V axis): Correct – Wnt/β-catenin, BMP4, and Activin/Nodal are key signals for dorsal-ventral patterning.
B (Mammalian limb D/V axis): Correct – Engrailed, Wnt/β-catenin, and BMP help define dorsal vs. ventral limb structures.
While Dpp (a BMP homolog) is involved, FGF and Hh are not primary players in Drosophila dorsal-ventral oocyte patterning. Instead, EGFR signaling (via Gurken) is key here.
Shh and FGF are crucial for A/P limb patterning (Shh from the zone of polarizing activity), Notch is not a major player in this axis.
The mRNA of the E.coli lac operon contains the open reading frames for lacZ, lacY, and lacA genes from a single cistron. It is observed that lacZ is translated more frequently than /lacY or lacA. Which one of the following statements best describes the reason for this observation?
1. The Shine-Dalgarno sequence is present upstream of lacZ, but not lacY or
lacA, affecting translation initiation rates.
2. Inhibitory RNA structures are present upstream of the AUG codon of lacY and lacA, but not lacZ, affecting translation initiation rates.
3. Variations in the Shine-Dalgarno sequence upstream of lacZ, lacY, and lacA
have different affinities for the ribosome, affecting translation initiation rates.
4. Inhibitory RNA structures are present in the lacY and lacA coding sequence
but not lacZ, affecting translation elongation rates.
Answer: 3
Explanation :
Each gene in the polycistronic mRNA has its own Shine-Dalgarno (SD) sequence. lacZ has a stronger SD sequence, allowing better ribosome binding and higher translation initiation. In contrast, lacY and lacA have weaker SD sequences, resulting in lower translation.
CSIR NET Life Science Dec 2024 Answer Key (Parts C) – Complete Explanations! Part 3
Also read :
Also read :
CSIR NET Life Science Dec 2024 Answer Key (Parts C) – Complete Explanations! Part 7
CSIR NET Life Science Dec 2024 Answer Key (Parts C) – Complete Explanations! Part 5
CSIR NET Life Science Dec 2024 Answer Key (Parts C) – Complete Explanations! Part 2
CSIR NET Life Science Dec 2024 Answer Key (Parts C) – Complete Explanations! Part 1
CSIR NET Life Science Previous Years Question Papers and answer keys
CSIR NET Life Science Dec 2024 Answer Key (Parts C) – Complete Explanations! Part 6
CSIR NET Life Science Dec 2024 Answer Key (Parts C) – Complete Explanations! Part 5
CSIR NET Life Science Dec 2024 Answer Key (Parts C) – Complete Explanations! Part 4
CSIR NET Life Science Dec 2024 Answer Key (Parts C) – Complete Explanations! Part 3
CSIR NET Life Science Dec 2024 Answer Key (Parts C) – Complete Explanations! Part 2
CSIR NET Life Science Dec 2024 Answer Key (Parts C) – Complete Explanations! Part 1
CSIR NET Life Science Previous Years Question Papers and answer keys
Join SACHIN’S BIOLOGY on Instagram or Facebook to receive timely updates and important notes about exams directly on your mobile device. Connect with Mr. Sachin Chavan, the founder of Sachin’s Biology and author of biologywala.com, who holds an M.Sc., NET JRF (AIR 21), and GATE qualifications. With SACHIN’S BIOLOGY, you can have a direct conversation with a knowledgeable and experienced.
![[PDF] Genomics and its Applications: 5 Modern Applications 7 Genomics and its Applications: 5 Modern Applications](https://biologywala.com/wp-content/uploads/2023/04/Applications-of-genomics-520x245.png)

![[PDF] Rod Cells and Cone Cells Function Notes: 10 Key Functions 9 [PDF] Rod Cells and Cone Cells Function Notes: 10 Key Functions](https://biologywala.com/wp-content/uploads/2023/07/Rod-Cells-and-Cone-Cells-Function-Notes-10-Key-Functions-1-520x245.webp)