DNA Isolation methods: 10 Techniques in 3 simple steps
DNA isolation methods play a critical role in molecular biology research as they form the first step in every study. The discovery of DNA can be traced back to 1869 when Friedrich Miescher, a Swiss doctor, carried out the initial crude extraction of DNA and referred to it as “nuclein.” He discovered the substance while researching proteins from leukocytes and found that its properties were fundamentally different from proteins.
Today, DNA can be extracted from a wide range of sources including soil, plant, and animal tissue, insects, protozoa, bacteria, yeast, and clinical samples such as fine needle aspirates of bodily fluids, biopsy samples, forensic samples such as dried blood spots, buccal swabs, and fingerprints, and soil samples. The choice of isolation and extraction method will depend on the type of sample being analyzed and the downstream applications for which the DNA will be used.
The linear arrangement of polynucleotides in DNA allows it to store biological information. It is translated into mRNA transcriptionally and then translated into an amino acid sequence that establishes the protein’s three-dimensional structure, which in turn establishes its biological function. Thus, DNA is regarded as the blueprint for life and delivers the instructions needed to maintain cellular activity.
When pH is neutral and salt concentration is physiological, the majority of DNA is in the B-form, which is the traditional right-handed double helix structure. Due to the syn conformation of its deoxyribose sugar ring, A-form nucleic acid, which is present in RNA-DNA and RNA-RNA duplexes, is thicker and has its base pairs packed closer together. Z-DNA is found in modest amounts in the cell and differs from B-form in that it has a left-handed helical structure that is thinner and has more base pairs per turn.
Due to the alternate stacking of purine bases that maintain a syn conformation and pyrimidine bases that are rotated into the anti-conformation, its phosphodiester backbone has a zigzag pattern.
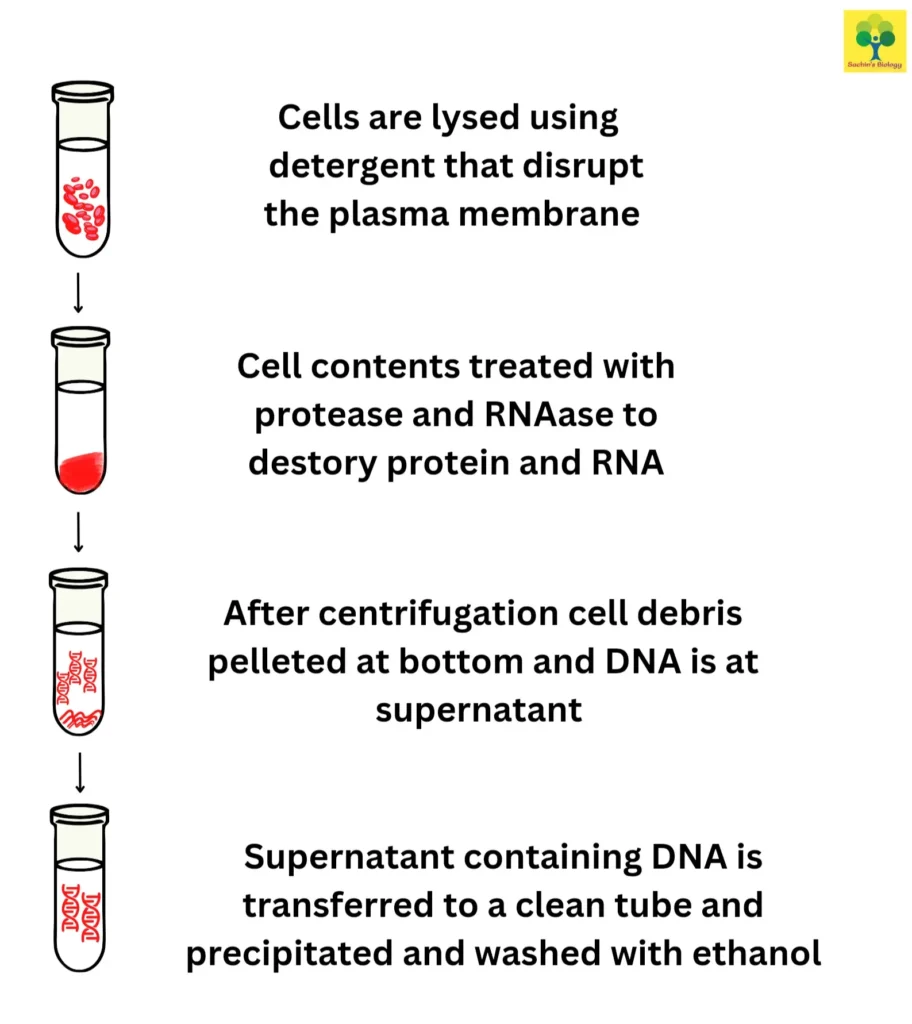
The three steps involved in the DNA extraction process are:
(1) rupturing the cytoplasmic and nuclear membranes;
(2) separating and purifying the DNA from other lysate-derived substances such lipids, proteins, and other nucleic acids; and
(3) concentrating and purifying the DNA. It is essential to guarantee the quality and quantity of the isolated DNA to carry out the intended downstream applications when selecting an appropriate method for DNA extraction. The time, cost, potential toxicity, yield, laboratory equipment and skill requirements, as well as the necessary sample size for the technique, should also be taken into account to optimise the DNA extraction method.
DNA isolation takes place by various techniques which are as follows:
1. Chromatography in DNA Isolation methods
Any type of biological material can be utilised to extract DNA using chromatography-based methods. This technique includes the Lathe and Ruthven-developed size exclusion chromatography (SEC), the Peterson and Sober-developed ion-exchange chromatography (IEC), and the Cuatrecasas & Wilcheck-reported affinity chromatography (AC).
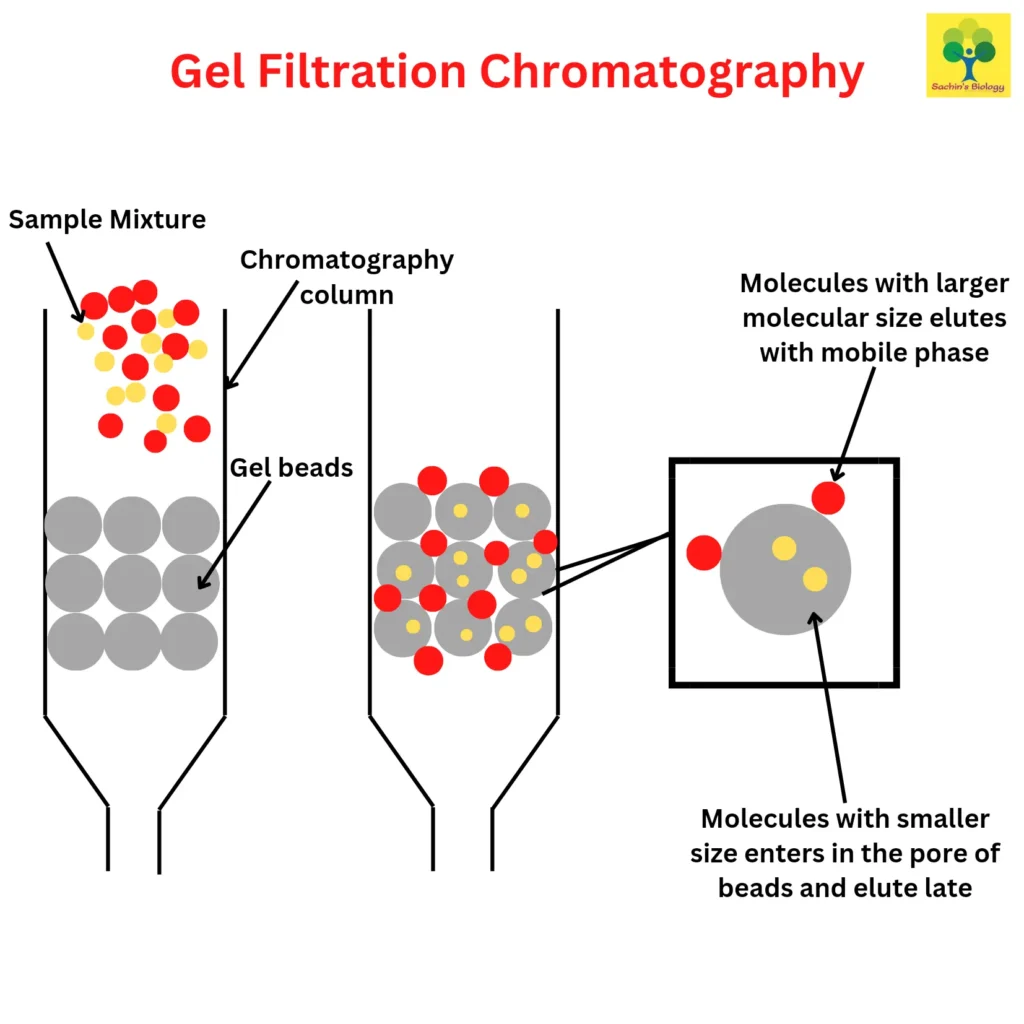
Molecules are separated using size exclusion chromatography (SEC), which takes into account both the size and form of the molecules. In contrast to gel permeation chromatography, which uses an organic solvent, the phrase “gel-filtration chromatography” is used when an aqueous solution is employed to move the DNA-containing sample through the chromatography column. Porous beads made of polyacrylamide, dextran, or agarose are present in the column.
Smaller molecules like mRNA and proteins enter the beads’ tiny pores and channels when the sample is put on top and passes through the column, however, DNA is prevented from doing so and escapes the matrix with its larger hydrodynamic volume. Since DNA is larger than the smaller molecules, it elutes from the column more quickly. SEC can be used on materials that are sensitive to pH changes and metal ion concentrations.
Ion-exchange chromatography is another chromatography-based technique for extracting DNA (IEC). A solution containing DNA anion-exchange resin is used to bind DNA with its positively charged diethylaminoethyl cellulose (DEAE) group in order to initially equilibrate the column. Other biological elements such as proteins, lipids, carbohydrates, metabolites, and RNA are eluted with medium-salt buffers while DNA is kept in the column.
The pH can then be brought down or high-salt buffers can be used to recover DNA. When compared to other DNA extraction techniques that produce high-quality DNA, including CsCl-gradient centrifugation, this technique is comparatively easy to execute.
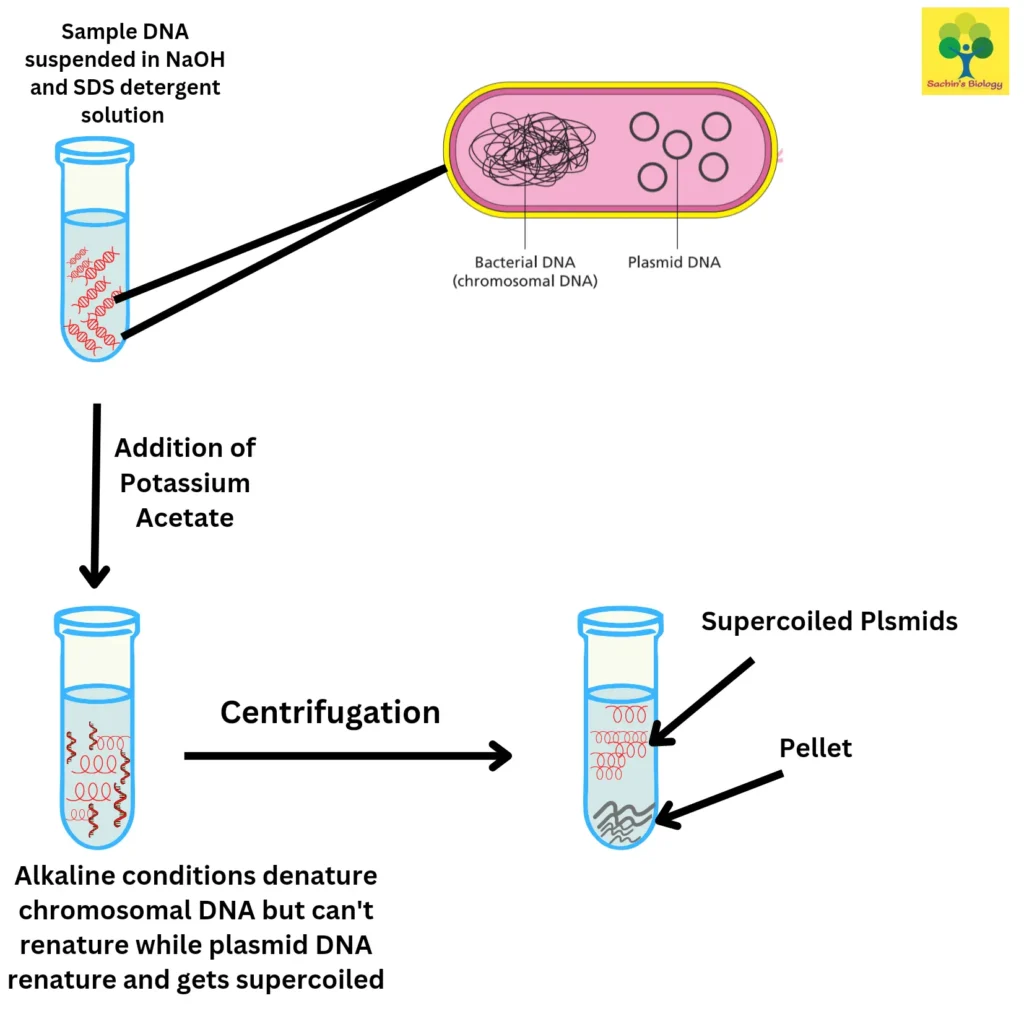
2. Alkaline extraction method
The technique, which was first presented in 1979 by Birnboim and Doly, is mostly used to harvest plasmid DNA from bacterial cells. For cell membrane lysis and protein denaturation, the sample is first suspended in an alkaline solution comprising NaOH and SDS detergent.
Chromosome DNA, which has a higher molecular weight than the intact, double-stranded plasmid DNA, is selectively denatured by the alkaline environment. Then, to neutralise the solution, potassium acetate is added. This causes the chromosomal DNA to denature and precipitate from the solution. After centrifugation, plasmid DNA in the supernatant can be obtained. The potential for plasmid DNA to become contaminated with chromosomal DNA fragments is a drawback of this approach.
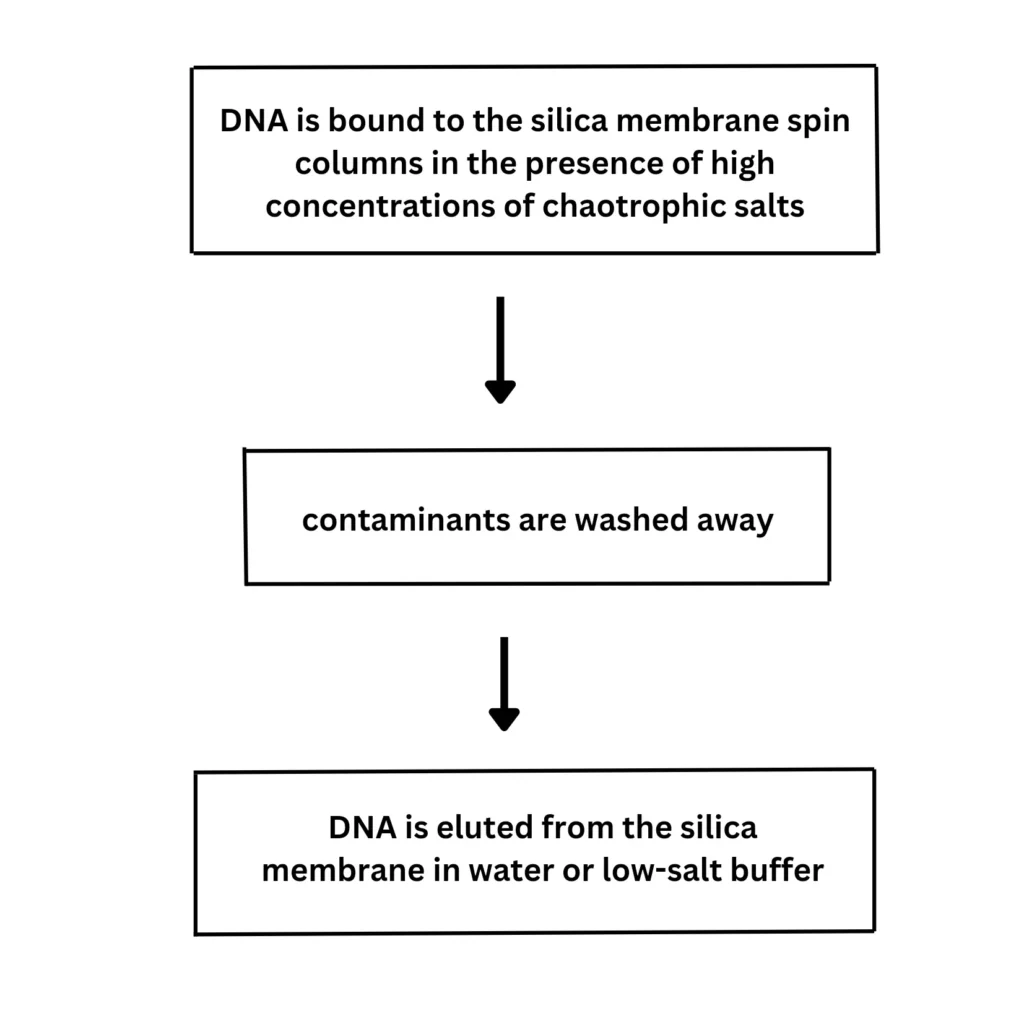
3. Matrices of Silica
In 1979, Vogelstein and Gillespie made the first mention of the silicates’ strong affinity for DNA. This DNA Isolation method is based on the idea that positively charged ions on the surface of silica can selectively attach to negatively charged DNA. The remaining cellular impurities can be removed using distilled water or a buffer like Tris-EDTA before the extracted DNA is eluted from the silica particles because the DNA is strongly linked to the silica matrix.
The use of a hydrated silica matrix for DNA extraction is just one example of the several modified protocols of this approach that have been published in the literature. The silica matrices approach is easy to use, quick to complete, economical, yields high-quality DNA, and can be automated. Due to their diminished ability to bind to DNA particles and continued attachment to them even after elution, silica matrices cannot be reused like the resins used in the anion exchange approach. The silica matrices can now be used several times thanks to newer techniques like maxXbond.
4. Salting-out technique
Miller, Dykes, and Polesky introduced the non-toxic salting-out method for DNA Isolation in 1988. The DNA-containing sample is combined with 3mL of proteinase K, SDS, and lysis buffer (0.4 M NaCl, 10 mM Tris-HCl, pH 8.0, and 2 mM EDTA, pH 8.0). Overnight, the mixture is incubated at 55 to 65 °C. After adding 6M of saturated NaCl, the liquid is agitated for 15 seconds before being centrifuged for 15 minutes at 2500 rpm. Protein precipitates because of the low solubility caused by the high salt concentration. Using ethanol, the DNA-containing supernatant can be precipitated after being pipetted into a new tube.
According to reports, the salting-out approach produces high-quality DNA that is equivalent to that obtained using the phenol-chloroform method, but it is preferable to that process since it uses less time, costs less money, and most importantly, the reagents are nontoxic. Additionally, DNA can be extracted from tissue homogenate, suspension cultures, and blood.
5. Extraction of Cetyltrimethylammonium Bromide (CTAB)
Doyle et al. published the CTAB DNA Isolation method in 1990. This technique involves adding DNA-containing samples to 2% CTAB at an alkaline pH. The extraction buffer precipitates acidic polysaccharides and DNA from the remaining cellular components in a solution with low ionic strength. The DNA from the acidic polysaccharides that precipitate with CTAB is then removed using solutions with high salt concentrations. Because of this, this approach is very helpful for extracting DNA from bacteria and plants that produce large amounts of polysaccharides.
Then, DNA is cleaned up using a variety of organic alcohols and solvents, including harmful substances like phenol and chloroform. The usage of hazardous chemicals and the length of time required for this approach are two of its main drawbacks.
6. DNA Isolation method of phenol-chloroform
The introduction of the phenol-chloroform DNA extraction technique is done by Barker et al. in 1998. To disintegrate cell membranes and the nuclear envelope, cells are first treated with a lysis buffer containing detergents such as sodium dodecyl sulphate (SDS). The lysis buffer may also contain 10 mM Tris, 1 mM EDTA, and 0.1 M NaCl. Then, 25:24:1 of the phenol-chloroform-isoamyl alcohol (PCIA) reagent is added. Isoamyl alcohol hinders emulsification, which helps with DNA precipitation while SDS and phenol both effectively denature proteins.
If the DNA molecules to be extracted are smaller than 10 kb, the mixture is combined to create a biphasic emulsion, and then they are vortexed, or if they are larger than 10 kb, they are gently shaken.
Upon centrifugation, the emulsion separates into two phases: the top aqueous phase, which is made up of DNA dissolved in water, and the bottom organic phase, which is made up of organic solvents and hydrophobic biological components like proteins. The organic phase can then be discarded after the aqueous part has been pipetted into a new tube. It is possible to prevent the aspiration of organic solvents by leaving a thin layer of the aqueous phase at the interface. Repeat these processes up until all protein has been removed from the aqueous-organic phase interface.
The industry standard for DNA extraction is the phenol-chloroform technique. It can be used to extract DNA from tissue homogenate, suspension culture, or blood. It has a significant yield and is
With a cost estimate of less than $5 for one sample, it is reasonably priced. The use of fume hoods is necessary since phenol and chloroform are poisonous, which is a significant drawback to this procedure. Extracted DNA samples have a greater purity than those made using other traditional techniques, but are not as high as those made using column-based techniques.
7. SDS Proteinase K
Ebeling et al. first identified Proteinase K as a serine protease in the fungus Engyodontium album in 1974. 20–50 L of proteinase K at 10–20 mg/mL are typically added for DNA extraction. Additionally, sodium dodecyl sulphate (SDS) is added to dissolve the nuclear envelope and cell membrane as well as to denature and unfold proteins so that proteinase K may use them as a protease. Using the phenol-chloroform or salting-out methods, the solution can be used to extract DNA after being incubated for 1–18 h at 50–60°C.
8. Using a silica column to extract DNA
In this procedure, the material is mixed with lysis buffer (0.05 M EDTA and 3 ml of 0.2 M Tris, pH 8.0), 1% SDS, and 100 micrograms of Proteinase K. The mixture is placed into a tube containing silica gel after an hour of incubation at 60°C. A 1:1 ratio of phenol and chloroform is added, and the mixture is forcefully mixed before being centrifuged at 1000 x g for five minutes. As a result, the DNA-containing aqueous phase is allowed to remain above the gel polymer layer, while the proteins and organic phase are trapped behind the silica column. The DNA-containing aqueous layer is then pipetted or decanted into a fresh tube, where it is dissolved in TE buffer.
The aspiration of organic solvents is typically prevented in the traditional method without silica gel by leaving behind a layer of the aqueous phase close to the interface to stop contamination.
However, using silica gel eliminates the need for an interface, improving the quality of the extracted DNA. Additionally, preventing direct contact with harmful chemicals, silica gel has been shown to produce 40% more DNA than the conventional organic solvent-based DNA Isolation method.
9. Magnetic beads
DNA purification and isolation using magnetic particles was the subject of a patent application by Trevor Hawkins in 1998. To bind DNA to its surface, magnetic nanoparticles can be coated with a DNA-binding antibody or polymer that has a particular affinity for DNA. Magnetic beads typically include a core made of magnetite or maghemite, and surface materials like silica and functional groups like sulphate and hydroxyl groups are also acceptable.
By creating a magnetic field at the bottom of the tube using an external magnet, it is possible to separate the DNA-bound magnetic beads from the cell lysate. The supernatant can then be removed by rinsing when the beads have accumulated at the tube’s bottom.
The ethanol precipitation DNA Isolation method can be used to elute the magnetic pellet, which is then incubated at 65°C to separate the magnetic particles from the DNA. The technique has been demonstrated to take less than 15 minutes to complete, which is substantially faster than other conventional methods that can take up to several hours. The DNA yield achieved by this method is comparable to that obtained in the other conventional methods.
Furthermore, because it is not dependent on centrifugation, it is perfect for automation and uses little equipment. The lack of shear forces, which could compromise the integrity of nucleic acids, is another benefit of this DNA Isolation method over centrifugation-based ones.
10. Extraction of Chelex-100
A technique for extracting DNA using chelex-100 was patented in 2011 by Xiong Hui, Xie Liqun, and Chen Jiayi. Chelex resin is a styrene-divinylbenzene copolymer that chelates metal ions with its iminodiacetic acid groups to serve as cofactors for DNases. Proteinase K, which is utilised to break down the DNases, and 5% chelex solution are added after overnight incubation. The residual membranes are subsequently lysed, and the proteins and DNA are denatured, by boiling the tissue sample in the 5% chelex solution.
Chelex stabilises the preparation by preventing DNA from being degraded by DNases that may remain after boiling. After centrifugation, the resultant single-stranded DNA can then be extracted from the supernatant. Since only one test tube is used and there aren’t many steps requiring manipulation, this method has fewer risks of contamination and incorrect pipetting.
The lack of purification, however, makes this approach inefficient for getting rid of PCR inhibitors. The isolated DNA may be unstable and unsuitable for RFLP analysis, which is another drawback.
11. Filter paper-based extraction method
In 2017, Rui Shi and Dilip Panthee wrote about a technique for isolating DNA from plant sources using filter paper. With each well containing a disc of WhatmanTM filter paper, a spin plate consisting of a 96-well plate (V-bottom) with a hole (about 1 mm in diameter) drilled into the bottom of each well is employed (approximately 8 mm in diameter). Centrifugation is used to filter samples that have been treated with lysis buffers. This technique significantly lowers the price of the approach by using filter paper for the glass fibre filters included in commercial DNA extraction kits.
Since Friedrich Miescher’s original DNA extraction in 1869, scientists have made incredible strides toward developing extraction techniques that are more dependable, quicker, easier, and more cost-effective while also producing a greater yield. The development of our understanding of the human genome has been made possible by newer, more effective methods.
These DNA Isolation methods have also significantly influenced the emergence of other scientific fields, including gene editing and customized medicine. Due to their limitations in producing yields with the best purity and ease of use, it appears that no one method is currently suited to all settings of DNA extraction.
When DNA is replicated?
During cell division, cells replicate their DNA before dividing to ensure that each daughter the cell gets a copy of the genome. During the S – phase (synthesis phase) of the interphase DNA replication takes place before the cell enters the cell division and the double-stranded DNA molecule is copied into two identical DNA molecules.
What is DNA isolation?
DNA isolation is also known as DNA extraction is the process of separating/purifying DNA from the sample by using different physical and chemical DNA Isolation methods. The purpose of isolating the DNA is to study individual genes, sequence the entire genome, modify the DNA section, and more.
What are the methods of DNA isolation?
Extracting the DNA from the sample is done by Various physical and chemical methods like chromatography methods, Alkaline extraction methods, Matrices of silica, Salting out techniques, Extraction of CTAB, Phenyl-chloroform extraction, SDS Proteinase extraction, Silica column to extract DNA, Magnetic beads, Extraction by Chelex- 100, and Filter paper-based extraction.
Why DNA extraction is important?
The primary importance of extracting DNA is for studying the various causes behind genetic diseases and developing diagnostic drugs for them. It is also necessary for Forensic science, for determining the paternity, and manufacturing of pharmaceuticals like hormones and vaccine production, and detecting bacteria and viruses in the environment.
What are the steps in DNA isolation?
Extraction of DNA involves the following steps-(1) Rupturing the cytoplasmic and nuclear membranes.(2)Separating and purifying the DNA from other lysate-derived substances such lipids, proteins, and other nucleic acids. (3) Concentrating and purifying the DNA. It is essential to guarantee the quality and quantity of the isolated DNA to carry out the intended downstream applications when selecting an appropriate DNA Isolation method.
The time, cost, potential toxicity, yield, laboratory equipment and skill requirements, as well as the necessary sample size for the technique, should also be taken into account to optimise the DNA extraction method.
How to isolate DNA
DNA isolation is the procedure that entails rupturing cells to release the DNA. For instance, in the case of bacterial cells, the cell membrane can be damaged and the DNA released using a detergent and salt solution (such as SDS). Mechanical and enzymatic techniques are frequently employed to separate animal and plant cells. After the DNA has been released, contaminants including proteins must be eliminated.
A precipitating agent, such as alcohol (like ethanol or isopropanol), or salt is generally used to do this (such as ammonium acetate). While the impurities stay in the solution, the DNA will solidify as a pellet at the bottom. Following precipitation, the DNA is often further purified using column-based techniques.
If you want important notes and updates about exams on your mobile then you can join SACHIN’SBIOLOGY on Instagram or Facebook and can directly talk to the founder of Sachin’s Biology and Author of biologywala.com Mr Sachin Chavan M.Sc. NET JRF (AIR 21) GATE, MH-SET
![[PDF] Production of Transgenic Plant for Biotic and Abiotic Stress: 7-Step Process 7 Production of transgenic plant](https://biologywala.com/wp-content/uploads/2023/08/Production-of-transgenic-plant-2-520x245.webp)
