Arabinose Operon complete notes

arabinose operon diagram
What is arabinose operon?
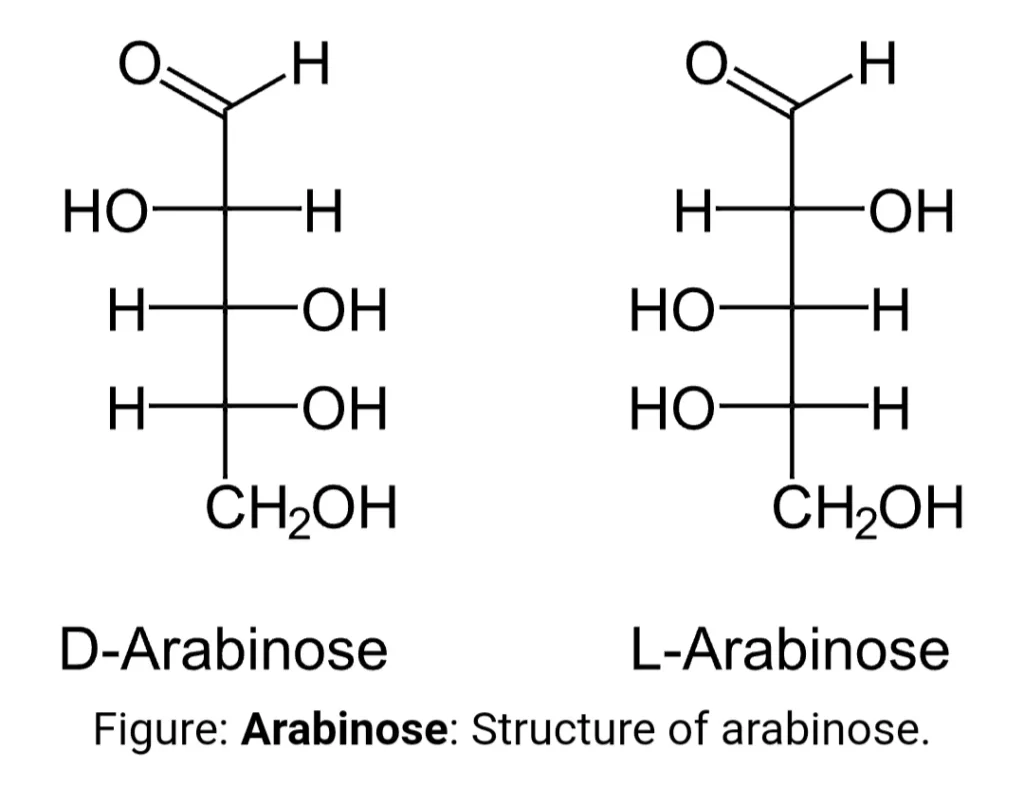
L-arabinose is also known as ara or ara BAD operon. Arabinose is a polysaccharide containing five carbon atoms with an aldehyde (CHO) functional group found in the cell walls of many plants and released in the human intestine after vegetables are eaten. This arabinose is not absorbed in the intestine, therefore, provides a source of carbon for bacteria. The L-arabinose operon contains the gene sequence that encodes the proteins needed for the catabolism of arabinose to xylulose 5- phosphate ( an intermediate of the pentose phosphate pathway) in the absence of glucose. This operon is found in prokaryotes like E.coli and is regulated both positively as well as negatively.
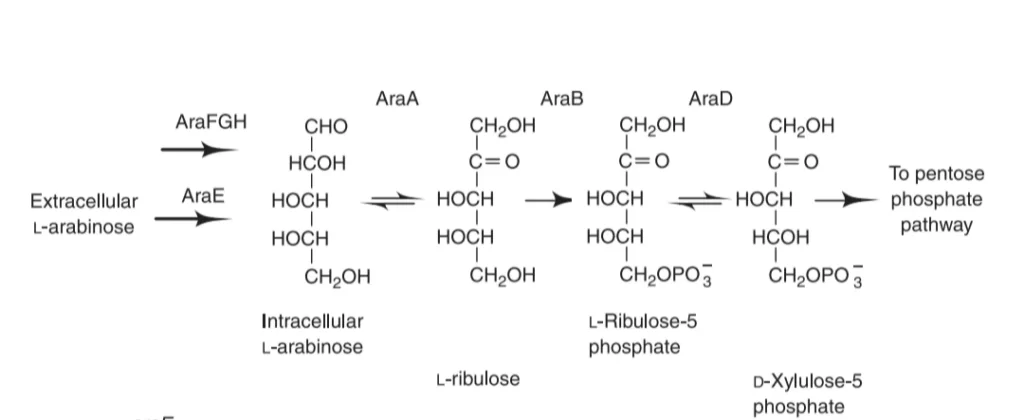
Different enzymes are required for the breakdown of arabinose to xylulose. These enzymes are synthesized by the specific genes present on the operon called as structural genes by the process of transcription and translation. But these genes are only made when arabinose is present. There are three types of structural genes present in the L-arabinose operon:ara B, ara A, ara D which are collectively called araBAD.
- Gene ara A encodes L- arabinose isomerase that converts arabinose to L-ribulose.
- Gene ara B encodes ribulokinase, which phosphorylates L-ribulose to form L-ribulose-5-phosphate.
- Gene ara D encodes L-ribulose-5-phosphate 4-epimerase that converts L-ribulose -phosphate to D-xylulose-phosphate.
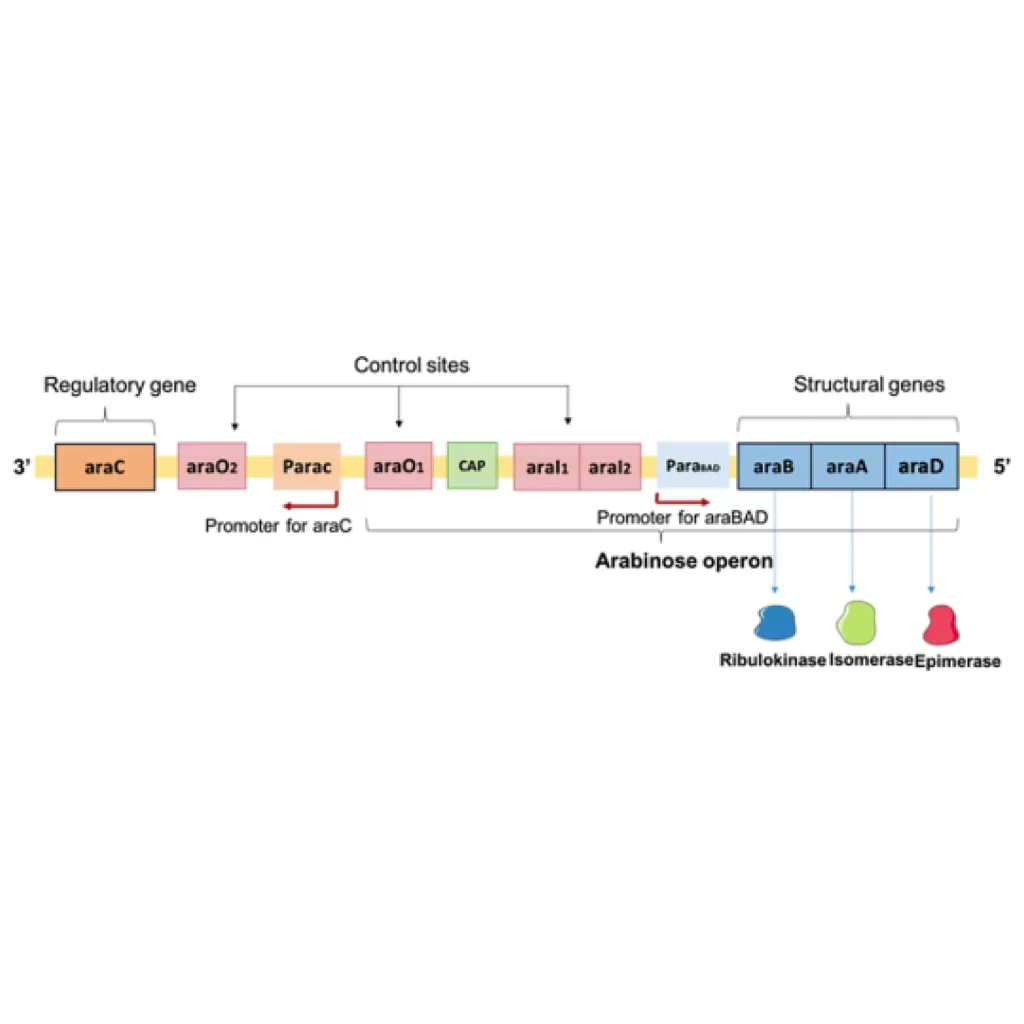
The promoter P araBAD controls the expression of these three structural genes( ara B, ara A, ara D ). ara C is the regulatory gene of arabinose operon which forms the regulatory protein araC and acts as a repressor. It can prevent transcription and also activates the transcription by binding to the inducer. Promoter P araC controls the expression of araC. The operators ara O1, ara O2, ara I1, and ara I2 act as a binding site for repressors. The ara I1 and ara I2 (I= Inducible sites) are DNA binding sites that when occupied by araC, induce expression.
Arabinose binds to the repressor AraC protein and helps RNA polymerase to initiate transcription. Catabolite Activator Protein (CAP) is the transcriptional activator. It is a homodimeric DNA binding protein that binds to the sequence that overlaps the 5’ side of the promoter. It helps in recruiting the RNA polymerase to the promoter site by binding to the C- terminal domain of RNA polymerase and increasing its affinity for the promoter sequence to overcome a weak promoter.
Ara C protein :
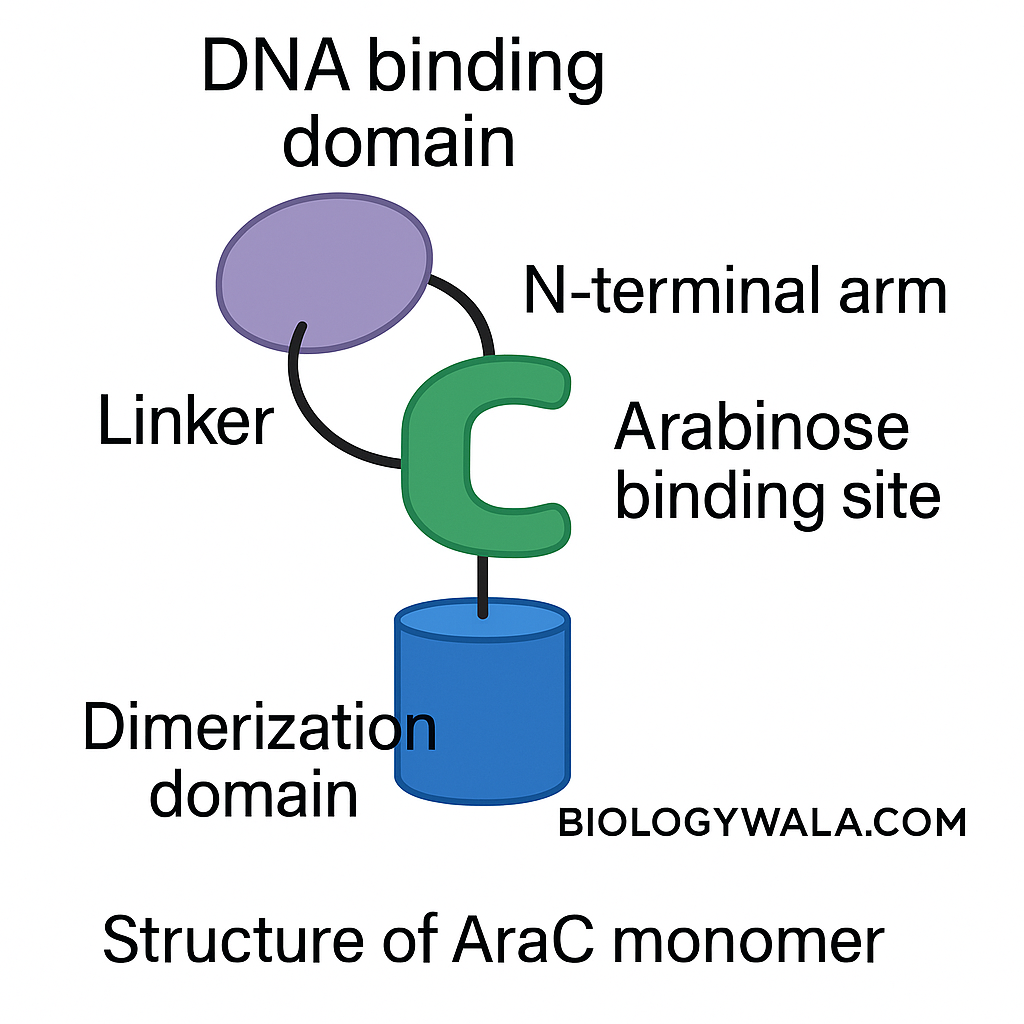
It is an arabinose binding protein that has an affinity for inducers and operators. The protein araC function as a dimer. It has two terminal domains: C- terminal domain and the N-terminal domain. The N-terminal domain is responsible for dimerization and C-terminal is responsible for the binding of DNA. The C- terminal domain face outwards and the linker region connects the N and C- terminal domains. The curvature in the N-terminal domain represents the pocket at which arabinose molecules can bind.
How does the arabinose get inside bacteria?
The ara operon itself does not involve any transport protein like the Lac operon. In the lac operon, the structural gene lac Y encodes for the transport protein permease. But in the case of ara operon, the transport proteins are independent of ara operon. Two independent operons control the transport of arabinose i.e. 1) ara E operon and 2)ara FGH operon.
The ara E encodes for the single protein which encodes for low-affinity H+ ion import and ara FGH encodes for three proteins called high-affinity ATP-dependent importers. Generally, the regulatory structure of ara E and ara FGH operon is very similar to the ara BAD operon. So the arabinose consumption depends on ara A, B, C, D, E, FGH genes where ara BAD controls the catabolism, ara C is the regulator, while ara E and ara FGH are the transport proteins of arabinose.
Negative regulation: Glucose(+) & Arabinose(-)
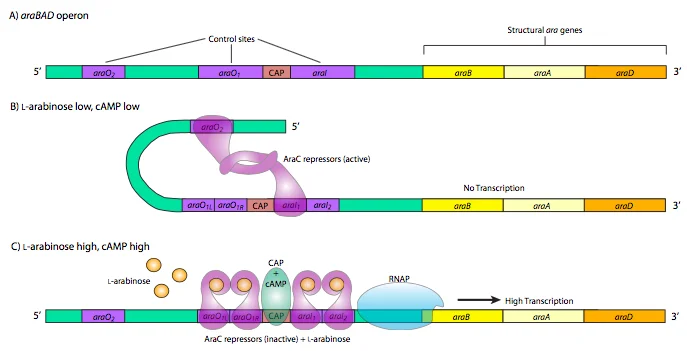
Glucose is the preferred source of energy in bacteria so when glucose is present in the cell and little or no arabinose is present, the ara C protein binds to the operator sequences ara O2 and ara I1. The two ara C proteins interact, which causes the DNA to loop around and prevents RNA polymerase from binding to the promoter and transcribing ara BAD.
Why ara C protein binds to the operator sequence O2 and I1 but not O1 and I2? The O1 site and I1 site are 50 bp apart from each other while on the other hand, the O2 site and I1 site are 210 bp away from each other, and I1 and I2 are only 4 bp away from each other. The I1 and I2 are too close to each other so ara C protein can’t bind to them because the ara C dimer is not flexible and can’t reach I2.
The reach of the ara C dimer has a lower limit cut off about (threshold) ~80 bp -100 bp. The distance between I1 and O1 is less than 50 bp which is under the threshold of 100 bp so ara C can’t reach O1 and if somehow ara C will reach the O1, the binding will be unstable. The only option available for ara C dimer for binding is O2 which is quite away and well above the threshold.
The ara C dimer can only bind to I1 and O2 when DNA forms a loop structure. The CAP site, O1, and promoter (P araC) of ara C are caught inside the DNA loop. The loop causes the DNA to supercoil which affects the topology of the DNA. This change in the topology of DNA causes a steric block so that the RNA polymerase can’t bind to the P araBAD and the transcription of the araBAD will be turned off.
Positive regulation: Glucose(-) & Arabinose(+)
When the arabinose is present for bacteria to consume naturally we would expect that the arabinose operon is activated which means the araBAD expression is upregulated. This regulation is driven by the change in the structure of the ara C protein. Typically, the ara C monomer has an N-terminal domain, C-terminal domain, and linker. When the arabinose is present in bacteria it can directly bind to the arabinose binding pocket of the N-terminal domain of ara C.
This binding causes a conformational change in the ara C monomer where the N-terminal domain arm flips out and acts like a lid for the arabinose binding pocket. This lid stabilizes the arabinose in the pocket.
In the default or non-active state, the DNA looping represses the araBAD expression but when arabinose is present it binds to the ara C dimer and causes the conformational changes in ara C. This changes the topology of the DNA by destabilizing the DNA loop. After the loop is destabilized the ara C binds to the I1 and I2.
In the off-state, the ara C dimer can only have long-distance contacts through DNA looping but when the arabinose is bound it changes the ara C dimer configuration so that it only forms short-range contacts(I1 & I2). In the upstream region of the inducer(I) there is an operator site O1. So the operator site can also have the ara C dimer bound to it. The arabinose bound to ara C at ara I sites interacts with RNA polymerase and with the help of CAP it promotes strong activation of araBAD expression.
Role of CAP in arabinose operon regulation:
Catabolite activator protein (CAP) is a small homodimeric DNA binding protein. It helps to recruit the RNA polymerase to the promoter site. If glucose is present in the cell the cAMP inside the cell decreases due to inactive adenylate cyclase. Enzyme adenylate cyclase synthesizes the cAMP (convert ATP into cAMP). This enzyme is negatively regulated by glucose transport.
So high glucose levels will block the function of adenylate cyclase and cAMP levels drop due to inactive adenylate cyclase and binding of cAMP to CAP site is not possible. In contrast, if the glucose levels are low, high levels of cAMP bind to the CAP site forming the cAMP-CAP complex and providing additional stimulation to the RNA polymerase and enhancing the efficiency of transcription.
Glucose(+) Arabinose(-)→ cAMP↓ → CAP inactive → Negative regulation
Glucose(-) Arabinose(+)→ cAMP↑ → CAP active →Positive regulation
This Arabinose operon is a unique example of an operon where the regulatory protein(ara C) acts as a repressor when arabinose is absent and as an activator when arabinose is present.
Hope you liked this article about Arabinose Operon. If you think there is something in this article or needs any kind of improvement you can let us know in the comment box below. After reading Arabinose Operon you will also like other articles on molecular biology.
You also like:
Control of gene expression in phages: Lytic cycle & Lysogenic cycle
Regulation of gene in Prokaryotes
Role of chromatin in gene expression and Gene silencing
[Download] Free Molecular Biology of Cancer PDF Book 4th Edi.
If you want important notes and updates about exams on your mobile then you can join SACHIN’SBIOLOGY on Instagram or Facebook and can directly talk to the founder of Sachin’s Biology and Author of biologywala.com Mr Sachin Chavan M.Sc. NET JRF (AIR 21) GATE!