Transcription in prokaryotes
Prokaryotic transcription:
Transcription in prokaryotes is the process in which a segment of DNA is copied into a newly synthesized strand of mRNA by the enzyme RNA polymerase. The transcription in prokaryotes takes place in the cytoplasm because they don’t have membrane-bound nuclei. The processes like transcription, translation, and mRNA degradation can occur simultaneously in prokaryotes because it does not have membrane-bound nuclei. The transcription and translation are coupled in prokaryotes and the translation of mRNA begins even before the completion of transcription. This is because mRNA is produced in the cytoplasm and the start codon of mRNA is available to ribosomes even before the entire synthesis of the mRNA molecule. The prokaryotic mRNAs are polycistronic means multiple genes are present on a single transcript and a single promoter controls the regulation of expression of all those genes.
The process of transcription in prokaryotes involves three steps: 1) Initiation 2) Elongation 3) Termination
Transcription initiation in prokaryotes:
Like any other biological process like DNA replication, transcription, or translation the initiation phase is the most important because in most cases this initiation phase is itself a rate-determining stage of all enzymatic reactions. The initiation phase involves enzymes and enzymatic reactions e.g. RNA polymerase enzyme in DNA transcription.
RNA polymerase in prokaryotes:
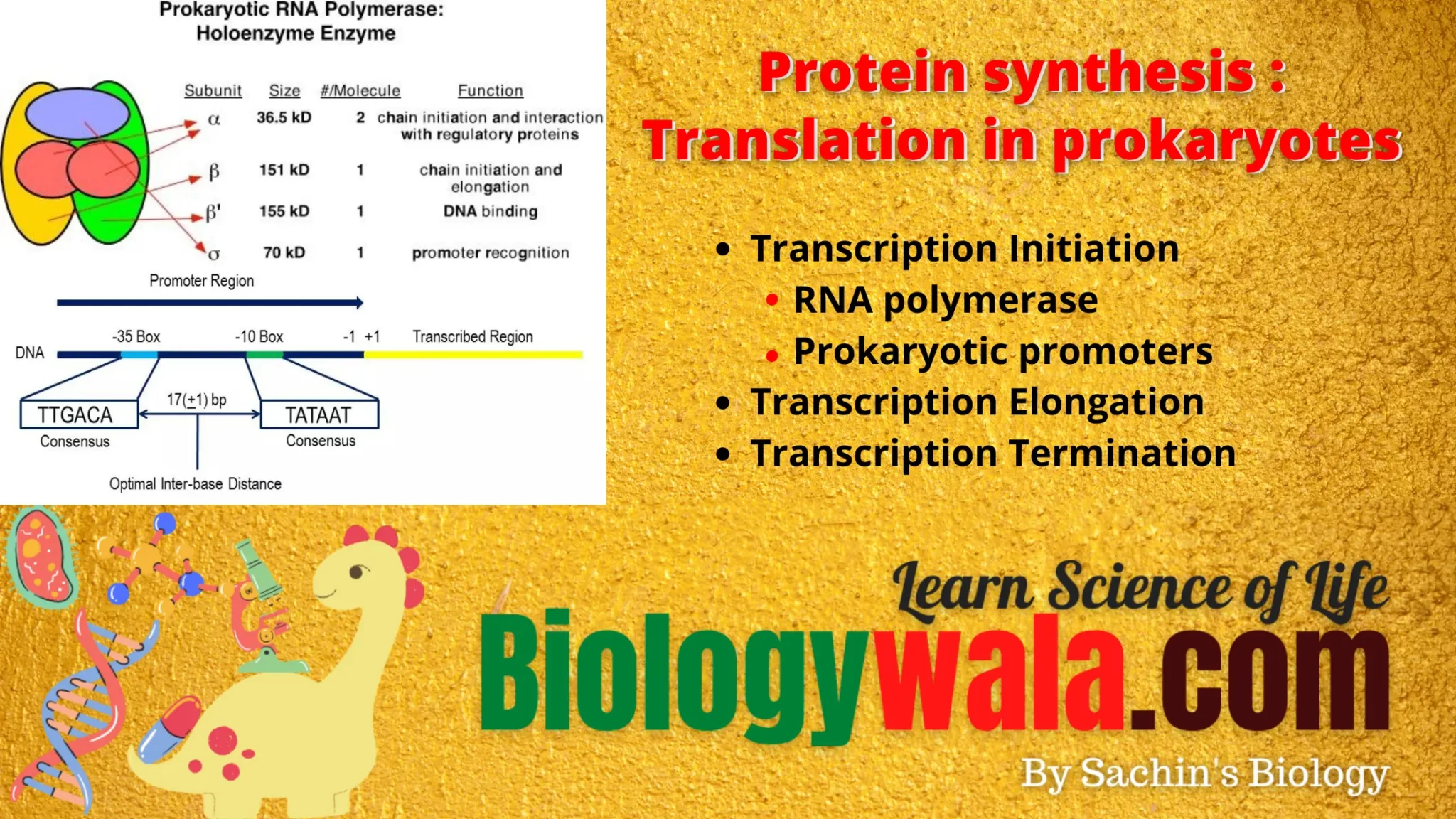
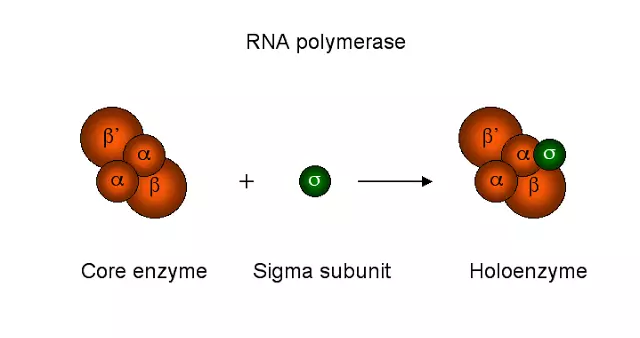
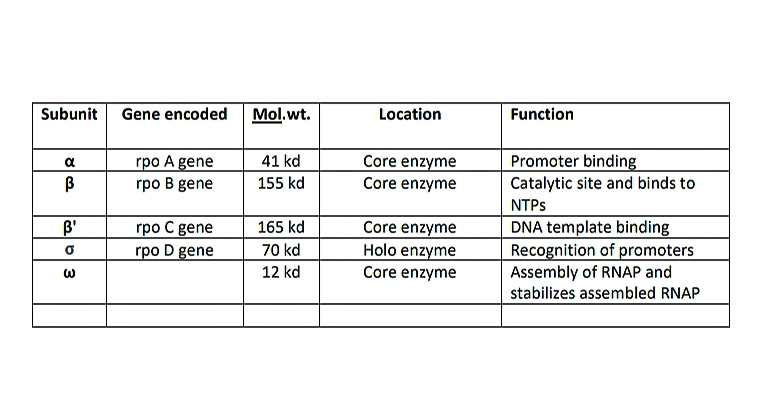
Prokaryotic RNA polymerase is a single type of RNA polymerase that can synthesize all the RNAs. It is a large multi-complex enzyme with a molecular weight of 465kd. The RNA polymerase contains 6 subunits and each subunit is encoded by a separate gene. The subunits are 2α, β, β’,ω, and σ (sigma). The five subunits 2α,β, β’,ω are together called core enzymes, and when the σ factor is added it is called Holoenzyme. Each subunit has a specific function.
1)2α subunit – helps in promoter binding 2)β subunit – It is a catalytic site and binds to NTPs 3)β’ subunit – DNA template binding 4)ω subunit – assemble the RNA polymerase and stabilizes it 5)σ subunit – recognition of promoters.
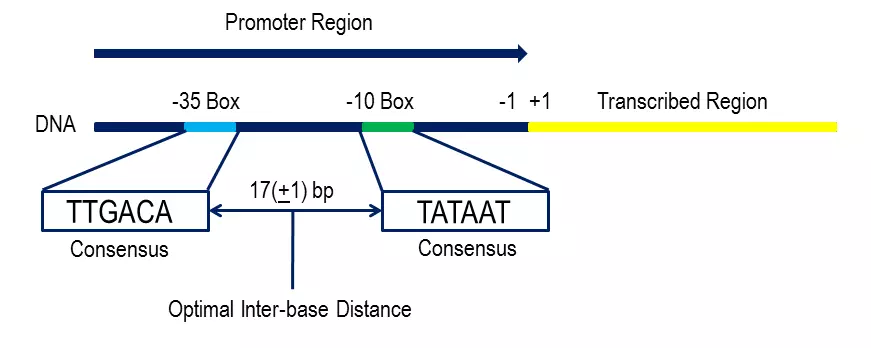
Prokaryotic promoters:
When a cell needs to build a protein, it recognizes the genes for proteins by their promoters. Promoters are the unique sequences of DNA that are located at the beginning of genes. In most cases, promoters exist upstream of the genes they regulate. The promoter sequence determines whether the corresponding gene is transcribed all the time, at one of the crucial times, or sometimes. The upstream of the initiation site, there are two promoter consensus sequences i.e. -10 region and the -35 region. These are the areas that are identical throughout all promoters and various bacterial species. The -10 consensus sequence is TATAAT(Pribnow box) and is located ten bases away from the transcription initiation site and the -35 sequence, TTGACA is located 35 bases away from the transcription start site. The -10 region is A-T rich and facilitates the unwinding of DNA templates.
The RNA polymerase starts binding to the DNA sequence but the binding is not firm because it is unable to detect the promoter sequence. So now the σ factor will come and help the polymerase to find the promoter sequence. When RNA polymerase along with the σ factor binds to the DNA sequence it stretches itself. This is because normally RNA polymerase along with the σ factor binds somewhere very much upstream of the start site even upstream of the promoter sequences. Thus, to find the promoter sequence RNA polymerase needs to stretch itself. After stretching, RNA polymerase finds the complete promoter sequence from -35 to -10. During the stretch, it can cover up to 60 to 80 bp.
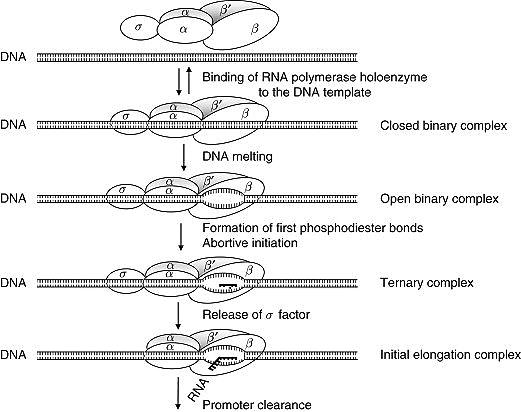
1)Once RNA polymerase along with the σ factor detects the promoter sequence by stretching itself, it forms a closed binary complex.
2)After that, DNA melting starts which mean RNA polymerase separates 10-14 bases extending from-11 to +3 and an open binary complex is formed. The change from closed binary complex to open binary complex is called isomerization.
3) RNA polymerase starts synthesizing nucleotides. When the RNA polymerase enzyme synthesizes the short RNA molecules of less than 10 bp it does not elongate further which is called abortive initiation. It is the repetitive synthesis and release of short-developing RNAs (less than 10 bp) by RNA polymerase. This happens because the σ factor lies in the middle of the RNA exit channelling the open complex. When the RNA polymerase manages to synthesize RNA more than 10 bp long, RNA is further elongated and the formation of a phosphodiester bond takes place between them, this complex is called the ternary complex (DNA+RNA+Enzyme). When the σ factor is attached to RNA polymerase, it can not start transcription. As the σ factor blocks the RNA exit channel and inhibits the transcription, the abortive initiation helps to remove the σ factor and initiate transcription. When the σ factor is released, after overcoming the abortive initiation, a long stretch of RNAs can be synthesized continuously up to the sequence of termination.
4) Transcription initiation will be completed when the sigma factor is removed followed by promoter clearance. Promoter clearance is the commitment of RNA polymerase enzymes to synthesize RNA. Thus RNA polymerase leaves the promoter to synthesize RNA.
Transcription elongation in prokaryotes:
The elongation refers to the process of the addition of nucleotides to the growing RNA chain. This is done by the enzyme RNA polymerase. It adds the nucleotides to form a long polymer of RNA. When the ribonucleotides are added they are in the phosphorylated form. According to the rule of complementarity, activated phosphorylated ribonucleotides lie to their complementary bases of the template strand. As these are in triphosphate form (ATP, GTP, CTP, UTP) they are converted into a monophosphate form. This is done with the help of the pyrophosphatase enzyme.
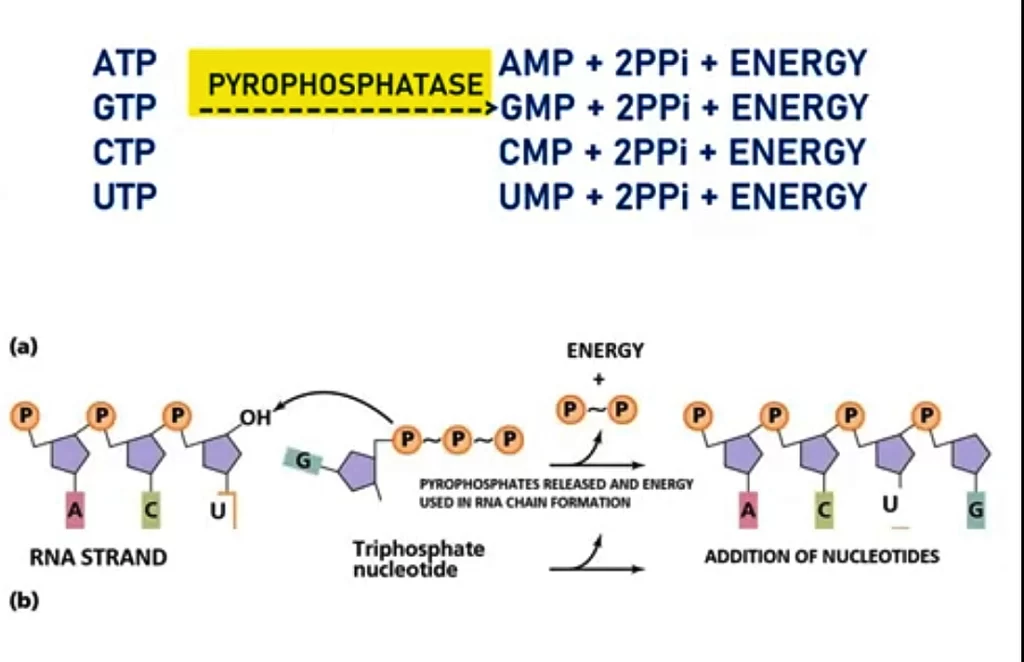
This enzyme converts the triphosphates ribonucleotides into monophosphate form with the release of pyrophosphate radicals. Pyrophosphate undergoes hydrolysis which liberates 2PPi ions and energy. This liberated energy is used in the chain formation and forming of temporary bonds between the base pairs of the template and newly transcribed RNA. RNA polymerase with the help of energy builds the phosphodiester bonds between adjacent nucleotides. This will continue until the RNA polymerase reaches the termination site. As the RNA polymerase moves on the template strand unwinding of DNA is done ahead and rewinding is done behind the RNA polymerase. The synthesis of mRNA is in the 5′ to 3′ direction of the growing strand at the rate of 40 nucleotides per second.
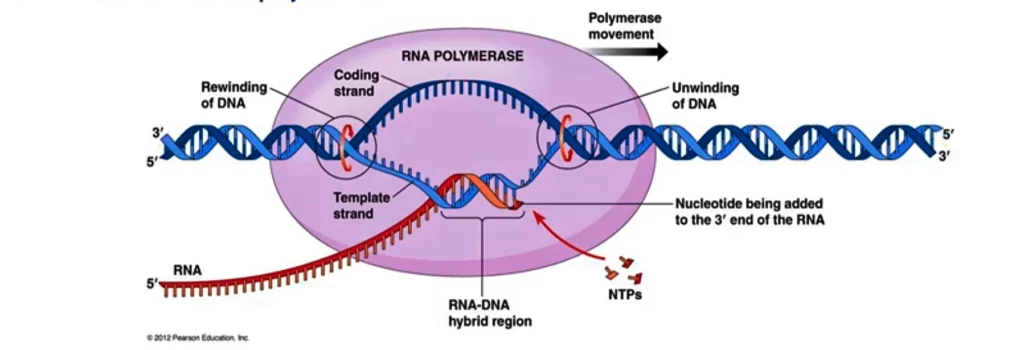
After the release of the σ factor, the protein NusA binds to the elongating RNA polymerase. It is associated with the RNA polymerase throughout the elongation and termination phase and helps in the termination and antitermination of transcription.
If any error or mutation occurs in the insertion of nucleotides, RNA polymerase also possesses the editing or proofreading activity but unlike DNA polymerase they have poor proofreading. Two types of proofreading are shown by RNA polymerase – Pyrophosphorolytic editing and Hydrolytic editing. When any wrong nucleotide is added to the template DNA the RNA polymerase will stop and proofread the transcript.
Pyrophosphorolytic editing– Once the incorrect nucleotide is added, the Pyrohosphorolytic editing is the very first way to rectify the mistake. In this type, a pyrophosphate (PPi) attack on the added wrong nucleotide. e.g. if AMP was the wrong nucleotide then PPi will convert the AMP into ATP. The incorrect nucleotides will be removed and RNA polymerase will add the correct nucleotides.
Hydrolytic editing – In this proofreading, proteins belonging to Gre and Nus protein family get activated. If an incorrect nucleotide is added RNA polymerase takes a halt and these proteins enter the active cleft of the core enzyme and remove nucleotides from nascent RNA and RNA polymerase will add the correct nucleotides again. In pyrophosphorolytic editing, the incorrect base pair is removed immediately while in hydrologic editing RNA polymerase has to fall back to fix the error.
Transcription termination in prokaryotes:
Termination refers to the ending of transcription. Why termination is important? In a genome, multiple genes are present in tandem so to prevent the expression of downstream genes the termination must happen at the end of the defined coding region. The termination occurs at the end of the coding region, usually called the termination branching point or kinetic window. Two main mechanisms are involved in the termination process – 1) Rho-dependent termination (Extrinsic termination) 2)Rho-independent termination (Intrinsic termination)
Rho-dependent termination:
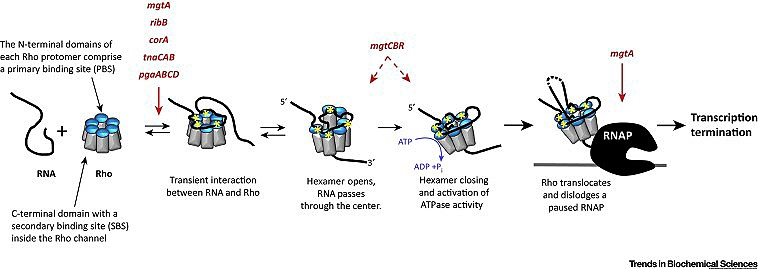
This termination mechanism requires the help of extra factors such as proteins that assist in the termination process. Most commonly this extra factor is the Rho protein or sometimes MFD protein. But the Rho factor is more commonly studied for the termination mechanism.
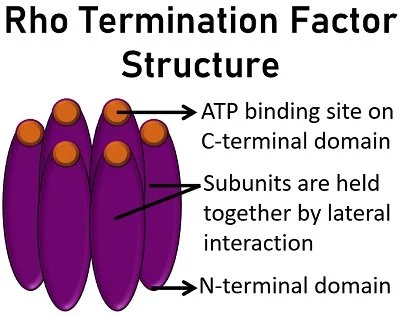
Rho protein is an RNA translocase that is a member of the Rec-A-family Helicase. It is a hexameric protein made up of six monomers which are in the form of a ring structure. Each terminal has an N-terminal domain and a C-terminal domain. In the ring structure, the N-terminal domains are positioned outwards and the C-terminal domains are inwards. The space in the centre is called the central pore. The N-terminal domain has the binding capacity for C-rich and G-poor RNA and it is also the primary binding site. The C-terminal domain has ATPase activity and it is the secondary binding site. The C-rich and G-poor primary binding sequences are also known as Rho-utilization sequences or rut sites (80-90 nt). The rut sites are located upstream of the termination site. These rut sites act as a signal for the binding of Rho protein. The recognition of rut sites happens when the enzyme has transcribed the rut sites. These sequences are present in the RNA. The mRNA moves to the central pore of the Rho protein and this initiates the secondary binding of the RNA by the C-terminal domain. Once the Rho protein is attached to the mRNA it will migrate toward the location of RNA polymerase in the 5′-3′ direction. The Rho protein reaches the elongation complex using ATPase-driven energy and hydrolyzes the RNA from the elongation complex by helicase activity. The interaction with Rho protein pulls DNA: RNA hybrid apart and releases the mRNA from the transcription bubble.
Rho-independent termination:
This termination mechanism is primarily driven by the RNA and DNA sequences. It is the information in DNA that gives rise to the RNA sequences that have the potential to form secondary structures (hairpin structure) in the transcribed RNA. The terminating sequence contains 20 base pair G-C rich rich regions (G-C dyad) followed by a short Poly-A tract and Poly-U tract respectively. The structure of the template DNA of the termination site contains Thymine followed by a G-C rich region which is then followed by a few Adenines. (3′ ~ATTT…….GCGCCATGGCCGC…..AAAT~5′) so the complementary sequence of RNA to the template DNA strand is 5’~ UAAA…….CGCGGUACCGGCG…..UUUA~3. The base pairing within the G-C rich region gives rise to the structure known as the terminator hairpin.
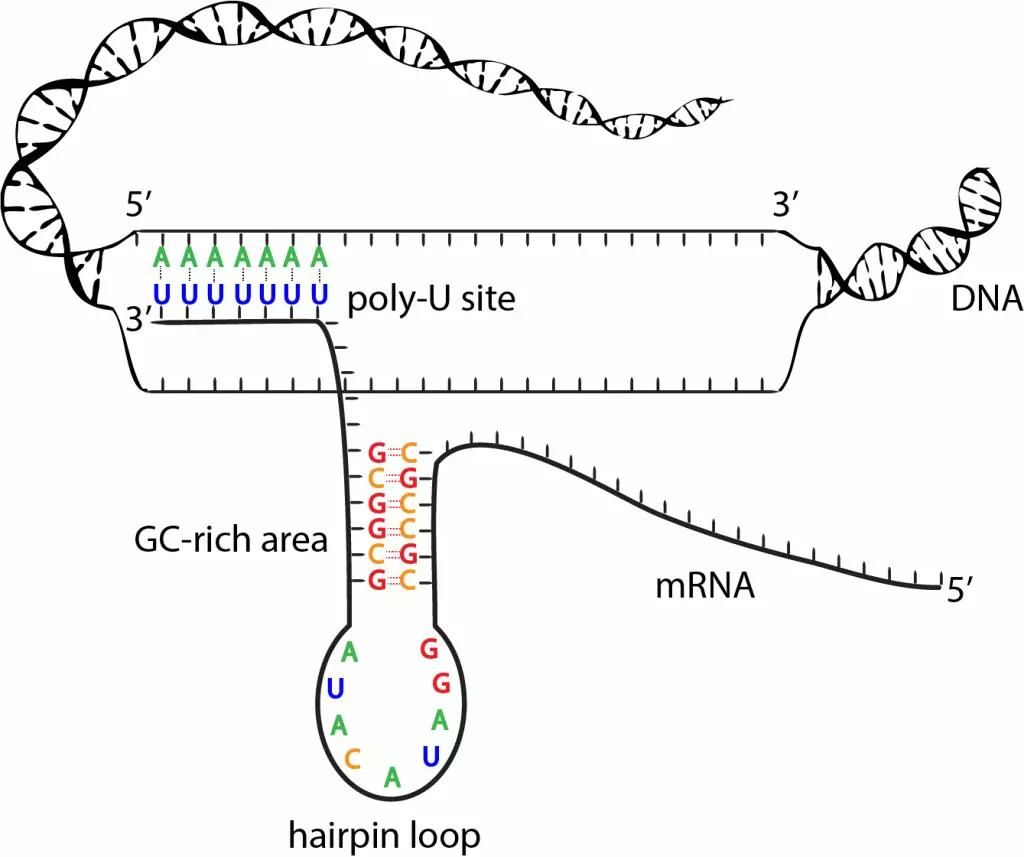
I) Elongation complex pause/ slow down:
The elongation complex travels on the coding region and for the termination, it needs to slow down. The pausing is initiated by the Poly-U tract as soon as it is transcribed at the transcription bubble. The elongation complex has a Poly-A tract hanging outside and the G-C rich region is in the RNA exit channel and not present as a hairpin structure yet. The Poly-U tract is present in the transcription bubble. The transcription of the Poly-U tract causes the elongation complex to slow down.
II)Formation of transcription hairpin:
The slow down of the elongation complex leads to the formation of a transcription hairpin. This initial stage of hairpin formation leads to the formation of a weak hairpin because in the hairpin structure Poly-A is at a 5′ end and Poly-U is at a 3′ end but the G-C rich region has some unpaired bases that can still be base-paired, therefore it is referred to as an incomplete or weak hairpin. The transition from a weak hairpin to a strong hairpin is largely a case of favourable energetics. Once the hairpin formation is complete the elongation complex is committed for the termination process. The complete hairpin formation makes the termination process irreversible. The complete hairpin loop has all the G and C regions paired.
III) Elongation complex disassembly:
There are two main models proposed for the disassembly of the elongation complex-
1)Hypertranslocation: In this model, the elongation complex just moves ahead without any addition of NTPs which shortens the RNA: DNA hybrid up to the several base pairs and thereby destabilizes the elongation complex.
2)Hybrid shearing: The hairpin extracts the newly made RNA by shearing RNA: DNA hybrid without a need for forwarding translocation.
Where does protein synthesis take place in prokaryotes?
The process of protein synthesis in prokaryotes involves transcription and translation. Both of these processes occur in the cytoplasm because the prokaryotic cell lacks a well-defined nucleus and membrane-bound cell organelles. The actual site for protein synthesis is in ribosomes but because of the lower level of cell compartmentalization ribosomes are scattered in the cytoplasm. So it is correct to say that the process of protein synthesis in prokaryotes takes place in the cytoplasm.
Difference between protein synthesis in prokaryotes and eukaryotes.
1) Eukaryotes have well-defined nuclei while prokaryotes lack a well-defined nucleus and membrane-bound organelles. So the process of protein synthesis in prokaryotes occurs in the cytoplasm whereas in eukaryotes transcription occurs in the nucleus and translation in the cytoplasm.
2) Protein synthesis in prokaryotes occurs even before the completion of transcription because mRNA is produced in the cytoplasm and the start codons of mRNA are available to ribosomes even before the entire synthesis of mRNA. In eukaryotes, protein synthesis(translation) occurs in the cytoplasm after the completion of transcription.
3) There are no introns present in the prokaryotic mRNA so the splicing of mRNA is not required whereas the splicing of mRNA takes place in the protein synthesis in eukaryotes.
4) Protein synthesis in prokaryotes can be fast as transcription and translation occur simultaneously. Eukaryotic synthesis is slower but they can check the mRNA before it is transcribed into the protein.
5) The 30s and 50s ribosomes are involved as well as capping and tailing do not take place in the prokaryotic protein synthesis. In eukaryotes, the 40s, and 60s ribosomes are involved and capping and tailing also take place during protein synthesis.
How is the transcription differs in prokaryotes?
1)The major difference between the transcription in prokaryotes and eukaryotes is that the transcription in prokaryotes occurs in the cytoplasm whereas in eukaryotes it occurs inside the nucleus. 2)A small amount of mRNA processing is required in prokaryotic transcription but in eukaryotic transcription extensive mRNA processing like the removal of introns and addition of exons, the addition of 5 caps, and poly-A tails takes place.3) A single RNA polymerase synthesizes all types of RNA in prokaryotes while eukaryotic transcription involves three types of RNA polymerase(I, II, III). 4)The transcription and translation are coupled in prokaryotes which means occur simultaneously but the coupled transcription and translations are not possible in eukaryotes as DNA is first transcribed in the nucleus and then transferred to the cytoplasm for the translation.
How does transcription end in prokaryotes?
The termination in the prokaryotic transcription can be Rho-dependent and Rho-independent. In the Rho-dependent termination mechanism, the protein Rho binds to the ribosome-free mRNA and translocates to the RNA polymerase. By the activity of the helicase, the Rho protein unwinds the RNA-DNA complex and mRNA is being released which marks the termination of transcription. In the Rho-independent mechanism, the information in the DNA gives rise to RNA sequence which has the potential to form secondary structures(hairpin loops) in transcribed RNA. The formation of hairpin loops causes the separation of the mRNA and RNA polymerase and mRNA is being released.
Hope you liked this article about Transcription in Prokaryotes. If you think there is something in this article that needs any kind of improvement you can let us know in the comment box below. After reading the prokaryotic transcription you will also like other articles on molecular biology.
You also like:
Control of gene expression in phages: Lytic cycle & Lysogenic cycle
Regulation of genes in Prokaryotes
Role of chromatin in gene expression and Gene silencing
[Download] Free Molecular Biology of Cancer PDF Book 4th Edi.
If you want important notes and updates about exams on your mobile then you can join SACHIN’SBIOLOGY on Instagram or Facebook and can directly talk to the founder of Sachin’s Biology and Author of biologywala.com Mr Sachin Chavan M.Sc. NET JRF (AIR 21) GATE!
![FREE [Download] Campbell Biology Concepts & Connections 9th Edition pdf book](https://biologywala.com/wp-content/themes/hueman/assets/front/img/thumb-medium-empty.png)

![[Download] Top 5 Books for Molecular Biology: Basic to Advance 12 Top 5 Books for Molecular Biology , molecular biology pdf books](https://biologywala.com/wp-content/uploads/2021/04/20210429_110135_0000-520x245.jpg)
