Control of gene expression in eukaryotes: 2 levels of expression
The control of gene expression in eukaryotes is more complicated than the gene expression in prokaryotes. The pathway for a gene to protein is much more complex in eukaryotes than the pathway in prokaryotes. This creates more opportunities for the control /regulation of gene expression in eukaryotes. When a gene product (RNA or protein) is present in a cell then it is said to be the expression of the gene, so any step in the pathway from gene to protein represents an opportunity for control of gene expression.
What exactly is the control of gene expression mean?
Control of gene expression in all about cells choosing which genes to use and which not to use. In a complete set of genes, cells only use certain genes at moment, depending on signals or changes in the environment.
- For example, When a single cell under goes cell cycle and divides by mitosis to give rise to the trillions of cells in your body, some of your cells become skin cells that protect you from microorganisms, or they became heart cells that contract to keep your blood flowing. Skin cells and heart cells would waste energy and resources if they made every protein all the time instead, these cells just use the genes they need, as they turn on specific genes and turn off rest.
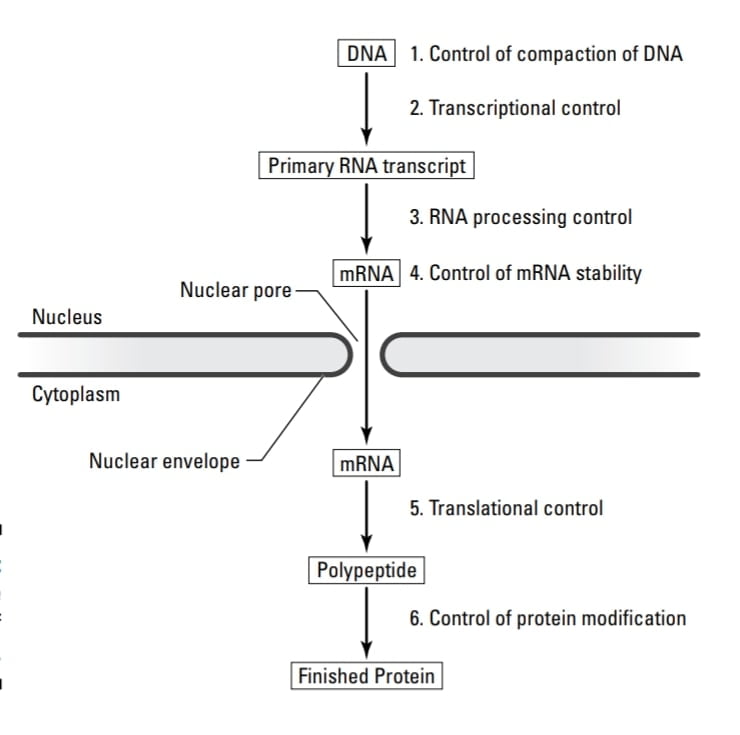
Levels of control of gene expression in eukaryotes:
There are two levels of control of gene expression in eukaryotes firstly Gene expression at the level of transcription and Post-transcriptional expression of the gene. These two levels of control of gene expression in eukaryotes govern part of the central dogma.
1. Gene expression at the level of transcription :
Control of gene expression in eukaryotes at the level of transcription gives clear information about the holistic process of gene expression in eukaryotes.
- Access to genes which are responsible for the transcription(i.e transcription factor) are affected by the compaction of DNA into chromosomes.
- The tight coiling of chromatin structure limits the access of these transcription factors.
- Cells contain a lot of DNA, if you took the DNA out of just one cell, it would be about 2 to 3 meters long. To fit this DNA inside, cells do a little creative packaging called DNA packaging.
DNA packaging in eukaryotes:
- Eukaryotes solve the DNA packaging problem by wrapping their DNA around the histone proteins. ( Most prokaryotes, do not have histones except domain Archea. So, in prokaryotes, DNA is supercoiled and compacted by nucleiod- associated proteins (NAPs).
- In Eukaryotic cells, DNA is packed into a chromatin structure. Chromatin is a network of DNA wrapped around histone proteins. (H2A, H2B, H3 & H4)
To pack the DNA into chromatin, cells do the following process:
- The possitively charged Histons are atracted by the negetively charged DNA. When DNA is wrapped around these clusters of histones, it looks like beads on a string. These beads are nothing but DNA plus histones called a nucleosome.

2. DNA and the string of nucleosomes twist until they condense in the form of coiled fibres. These fibres are called 30-nanometer fibres as they are 30- nanometer wide.
3. Continued twisting packs the DNA more tightly until DNA is very condensed. For example, imagine a rubber band and keep twisting it in one direction until it begins to form knots. Same in the case of DNA!
- The expression of the gene is controlled on the level of DNA packaging by some epigenetic modification.
- Epigenetic modifications are responsible for the regulation of gene expression in cells. Epigenetics deals with the study of process of cells which controls gene activity without changing the DNA sequence. This modification to DNA regulates whether genes are turned on or off.
- Chromatin remodeling is highly implicated in epigenetics. Transcriptional regulations involve active interaction between transcription factors and chromatin. Chromatin remodeling is the rearrangement of chromatin from a condensed state to a transcriptionally available or accessible state, which allows transcription factors to access DNA and control gene expression. In general, the lesser the condensation of chromatin, the more it is easy for transcription factors and other DNA binding proteins to access DNA and perform their duties.
( Heterochromatin – chromatin is highly packed and not actively transcribed.
Euchromatin – chromatin is loosely packed and accessible for transcription.)
Chromatin remodelling is a non-covalent modification.
The alternation or rearrangement in chromatin is done by following epigenetic modifications resulting in transcriptional activation or repression.
- DNA methylation – It is the addition of methyl (CH3) groups to DNA often modifying the function of genes.DNA methylation process is a covalent addition of methyl group at the 5-carbon of the cytosine ring resulting in 5-methylcytosine, called DNA methyltransferases (DNMTs).

- Most CpG sites are heavily methylated. CpG site is a location within a DNA sequence in which cytosine and guanine appear consecutively. (5’C- P- G -P -C -P – G 3′). When a CpG island (sites of CpG cluster ) in a promoter region is methylated certain protein binds to it which blocks the binding of transcription factors and inhibit the transcription.
- Histone modification: Histone acetylation and deacetylation are essential parts of gene regulation. In histone acetylation, the acetyl group is added usually to lysine residues at the N-terminus, by histone acetyltransferase (HATs). HAT proteins attach the acetyl group to histone proteins, which neutralize their positive charge. DNA is less attracted to histones when they (histones) lose their positive charge and the bond between DNA and histones is weakened. DNA decondensed due to weakened bonds and allows transcription factors to bind and initiate transcription. While a histone deacetylation acetyl group is removed from lysine by histone deacetylase (HDACs) which allow histones to wrap DNA more tightly and block the transcription.

- Histone methylation of some lysine and arginine residues results in transcriptional activation. In this process, a methyl group is added to lysine or arginine by an enzyme histone methyltransferase (HMT) with the help of S- Adenesyl Methionine(Donor). Depending on the attachment of the methyl group, the activation or inactivation of transcription will be decided. If the methyl group is attached to the 4th and 36th positions of lysine transcription get activated and if the methyl group is attached to the 9th and 27th positions, the transcription gets inactive. It is a reversible process, so demethylation occurs by histone demethylase (HDM).

- Histone phosphorylation also plays an important role in transcription control. As this is a reversible process, the phosphate group is attached to histone by enzyme kinase with the help of ATP (Donor) called histone phosphorylation and is removed by the enzyme phosphatase called histone dephosphorylation. All these histone modifications (acetylation, methylation, and phosphorylation ) are covalent modifications.

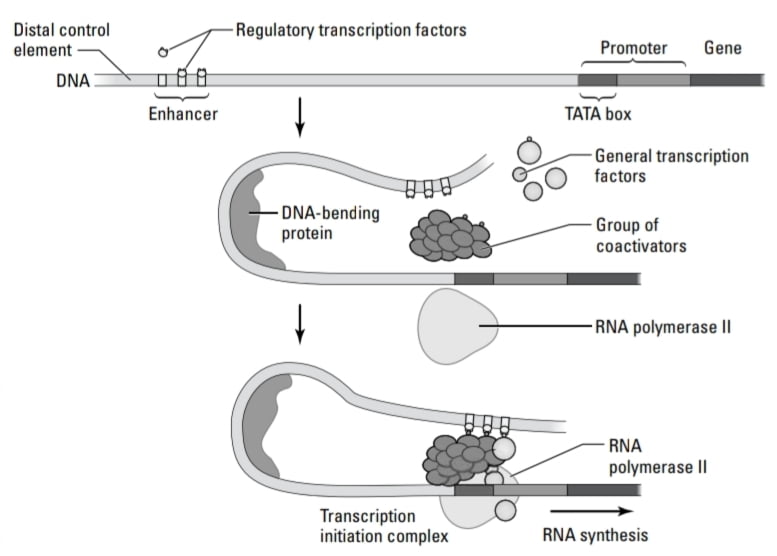
- Controlling transcription: Transcription in eukaryotes is controlled by DNA- binding proteins called regulatory proteins, which bind to regulatory sequences are more diverse in eukaryotes than prokaryotes.
- The promoter-proximal element is located close to the promoter and acts as regulatory sequences. Regulatory proteins bind to the promoter-proximal elements, transcription is turned on.
- Enhancers are cis-acting elements. (cis-acting elements – influence expression of adjacent genes on same DNA molecule. Trans-acting elements – diffuse through the cytoplasm and act at target DNA sites on any DNA molecule in the cell.) Enhancers are thousands of base pairs away from the initiation site and required for maximum transcription of a gene. They contain a short sequence of elements, activators bind to these sequences and form protein complexes. These protein complexes postulate to bring the enhancer complex close to the promoter and increase transcription. As enhancers are far away from promoters, how do they work? This is because of the possible looping of DNA. Enhancer shows deformability and bendability by which looping of DNA is possible and brings transcription factors together.

- Silencers (trans-acting elements)are the regulatory sequences that are also far away from genes they regulate but after binding to the regulatory protein transcription is blocked.
- The regulatory protein that binds to the regulatory sequence to turn on transcription is called the transcription factor. Two types of transcription factor present in eukaryotic cells:
- 1)Basal (general) transcription factor is required for transcription of any gene in all cell types. It is the part of the transcription initiation complex (a group of proteins that works together to attract RNA polymerase and helps promoters to bind).
- 2) Regulatory transcription factor is more targeted as it binds to a regulatory sequence of specific genes to turn them “on” or “off”.
- Coactivators are the proteins that affect transcription without binding to DNA. Coactivators bind to general and regulatory transcription factors and bring them together to form the transcription initiation complex.
- Transcription factors bind to a regulatory sequence to control gene expression in eukaryotes. To turn genes “on ” transcription factors bind to promoters and enhancers and to turn genes “off” transcription factors bind to silencers.
2. Post-transcriptional expression of the gene :
To understand the control of gene expression in eukaryotes, one must understand the concept of Post-transcriptional expression of the gene.
- mRNA processing can affect the conversion of a pre- mRNA into a finished mRNA molecule. It is regulated by a protein that interacts with spliceosomes. Through alternative splicing, spliceosomes can splice one type of pre-mRNA in different ways which leads to differences in proteins. Thus, alternative splicing produces several different gene products from one original pre-mRNA.

- The life span of mRNA or mRNA stability is controlled by RNA interference. Small RNA molecules micro RNA (miRNA) bind to complementary sequences on mRNA molecules making them for destruction by the cell.
- Translations of mRNA may be inhibited or increased by regulators. Regulating protein can bind to mRNA blocking their translation. The cell may be modifying the 5′ cap or poly-A tail of mRNA to block translation.
- Prevention of translation of mRNA inhibits the gene product from being made, blocking expression of the gene from which mRNA was made.
- After the completion of translations, protein is available in the cell. However, cells can still control the effect of that protein by regulating its activity.
- The activity of other proteins is often controlled by the addition or removal of a phosphate group. i.e. by the process of phosphorylation or dephosphorylation.
- All stages of gene expression can be regulated but the main control point for many genes is transcription.
Difference between regulation of gene expression in prokaryotes and eukaryotes : gene expression prokaryotes vs eukaryotes
- Gene regulation in prokaryotes suggests a simpler process. As prokaryotic organisms are single-celled with a lack of a well-developed nucleus; their DNA floats freely in the cytoplasm of the cell. Therefore, transcription and translation occur simultaneously in the cytoplasm, and the regulation of a gene occurs at the transcriptional level because when more protein is required more transcription occurs, and when it requires less there is less occurrence of transcription.
- While in Eukaryotes this process is more complex. In eukaryotes, the regulation during transcription and RNA processing takes place in the nucleus and translation takes place in the cytoplasm.
- Prokaryotic genes are likely to be regulated together in groups whereas each eukaryotic gene is regulated separately.
- In eukaryotes, three types of RNA polymerase are present(i.e. RNA polymerase I, RNA polymerase II & RNA polymerase III) while prokaryotes utilize one RNA polymerase for all transcription of types of RNA.
- The initiation of transcription is simpler in prokaryotes. In eukaryotes, it involves many transcription factors and RNA polymerase to form the transcription preinitiation complex. More steps are involved in mRNA processing like capping, splicing, poly-A tail addition.
- Another difference is eukaryotes express one gene at a time, they don’t express their genes all at once while prokaryotes express all the genes simultaneously.
- Splicing of introns is involved in eukaryotic gene regulation but in prokaryotes, this process is absent as they don’t contain introns.
What gene controls gene expression?
Regulatory genes control the expression of genes by producing regulatory proteins or transcription factors (i. e. activators, and repressors). These regulatory proteins activate or inhibit the transcription by binding to the DNA binding sites near the promoter region. Although the mechanism of control of gene expression is different in prokaryotes and eukaryotes.
What are 2 mechanisms by which eukaryotic gene expression can be regulated?
The process of regulation of gene expression in eukaryotes is complex which creates more opportunities for the regulation of gene expression in eukaryotes. The mechanism of gene regulation has two important levels i.e. regulation of gene at the level of transcription and post transcription. At the level of transcription, transcription factors bind to regulatory sequences(enhancer, silencer) to control gene expression. Post transcription mechanisms involve steps like mRNA processing(RNA splicing), mRNA stability, prevention of translation of mRNA to avoid the production of the gene product. Chromatin remodelling also plays role in the regulation of gene expression by altering the structure of chromatin. Some epigenetic changes are also involved like DNA methylation, histone modification in the process of gene expression control.
What prevents gene expression?
Regulatory proteins like repressors and activators control the gene expression in the cell. However, regulatory proteins like a repressor prevent or inhibit the expression of the gene. These regulatory proteins bind to the regulatory sequences(enhancer, promoter) and inhibit the progress of RNA polymerase along the DNA strand which results in the blocking of transcription.
Gene expression prokaryotes vs eukaryotes
1. Gene regulation in prokaryotes suggests a simpler process. As prokaryotic organisms are single-celled with a lack of a well-developed nucleus; their DNA floats freely in the cytoplasm of the cell. Therefore, transcription and translation occur simultaneously in the cytoplasm, and the regulation of a gene occurs at the transcriptional level because when more protein is required more transcription occurs, and when it requires less there is less occurrence of transcription.
2. While in Eukaryotes this process is more complex. In eukaryotes, the regulation during transcription and RNA processing takes place in the nucleus and translation takes place in the cytoplasm.
3. Prokaryotic genes are likely to be regulated together in groups whereas each eukaryotic gene is regulated separately.
4. In eukaryotes, three types of RNA polymerase are present(i.e. RNA polymerase I, RNA polymerase II & RNA polymerase III) while prokaryotes utilize one RNA polymerase for all transcription of types of RNA.
5. The initiation of transcription is simpler in prokaryotes. In eukaryotes, it involves many transcription factors and RNA polymerase to form the transcription preinitiation complex.
6. More steps are involved in mRNA processing like capping, splicing, poly-A tail addition.
7/Another difference is eukaryotes express one gene at a time, they don’t express their genes all at once while prokaryotes express all the genes simultaneously.
8.Splicing of introns is involved in eukaryotic gene regulation but in prokaryotes, this process is absent as they don’t contain introns.
Hope you liked this article about the Control of gene expression in eukaryotes. If you think there is something in this article or need any kind of improvement you can let us know in the comment box below.
You also like:
Regulation of gene in Prokaryotes
[Download] Free Molecular Biology of Cancer PDF Book 4th Edi.
If you want important notes and updates about exams on your mobile then you can join SACHIN’SBIOLOGY on Instagram or Facebook and can directly talk to the founder of Sachin’s Biology and Author of biologywala.com Mr Sachin Chavan M.Sc. NET JRF (AIR 21) GATE!


3 Responses
[…] control of gene expression in eukaryotes is completely different from the gene expression in prokaryotes. The pathways that produce gene is […]
[…] the article Role of chromatin in gene expression and Gene silencing, we will be discussing chromatin modifications, Gene silencing, Histone […]
[…] in which both lytic and lysogenic cycles are present is called temperate phage. The regulation of gene expression in phages is all about how the lytic cycle gets switched to the lysogenic cycle and vice-versa. […]