Download Regulation of gene in Prokaryotes complete pdf notes
Introduction: Regulation of genes in Prokaryotes
What is the regulation of gene expression?
Regulation of gene expression involves the mechanism that is used by cells to develop or restrict the production of specific gene products (protein or RNA ). When a gene leads to a formation of protein and plays a function i.e. phenotypic effect at molecular, biochemical level then it is said to be an expression of the gene.
Various factors control how much a gene is transcribed these are called Transcription Factors.
Activators:
i) It helps RNA polymerase to bind at the promoter site. ii) In eukaryotes, activators are important as eukaryotic genes have weak promotors while in prokaryotes, strong promoters are present where repressors are required to stop strong promoters. iii) Activators bring allosteric changes in RNA polymerase. iv) Transcriptional activators affect the chromatin structure surrounding the target gene by histone-modifying protein (HAT – Histone Acetyl Transferase, HMT – Histone Methyl Transferase ) and chromatin remodelling complexes.
Repressor:
i) Transcriptional repressors can bind to the promoter or enhancer region to block Transcription. But a little amount of gene can transfer at a basal level as a repressor and also have a half-life after which it gets removed. e.g -Lac I ,trpR,Cro,CI ,AraC. ii) Repressor protein brought negative allosteric changes leading to dissociation of RNA polymerase iii) Chromatin remodelling – Histone deacetylation protein ( HDAC ) removes acyl leading to binding of Histone to DNA and convert the DNA to heterochromatin.
Activator and Repressor proteins work in dimeric or trimeric form as a sequence of operators and enhancers are bipartite and palindromic sequences.
Each activator or inhibitor should have a DNA binding Domain along with a motif.
Motifs:
Motifs are short nucleotide sequences used to control the expression of genes by dictating under which condition a gene will be turned on /off.
Types of Motif:
- Helix – Turn – Helix motif – HTH is a major structural motif which binds DNA. It is the most common motif in prokaryotes. e.g. CAP ,DNA ,C1 ,Lac l ,AroC.
- Zinc finger motif – i) These are small structural protein motifs and it’s useful to maintain the structural fold of DNA because Zinc(Zn+) holds all the structures together.
Types of Zinc finger motif :
1)C2H2 motif – Two pairs of Cysteine of β-strand and two pairs of Histidine at C- terminus of α-helix bind to Zinc.
E.g. TFIIIC
2)C4 motif – It is structurally different from the C2H2 motif as Zinc twist binds DNA by two copies of the C4 motif.
- Leucine zipper motif – i) It is simply a dimeric protein i.e. two monomers with two α-helix. ii)In the α-helix every 7th residue is Leucine. Iii) Leucine is helping to zip two α-helix together by creating hydrophobic domains.
E.g. c-fos,c-jun,myc
d) Helix-loop-Helix – i) It helps in DNA binding and dimerization ii) There are Two α-helix present and approximately 15-16 residues .iii) Both the α-helix are differentiated by a loop and these loops are made up of 12 – 28 amino acids.
Regulation of gene in Prokaryotes: Lac Operon
The Regulation of genes in Prokaryotes occurs through inducible operons or repressible operons. Which have proteins that bind to activate or repress transcription depending on the environment and needs of the cell.
The lac operon : An Inducible Operon
Jacob and Monad studied lactose metabolism in E.coli and proposed the Lac Operon concept. Lac Operon is also called leaky Operon as transcription is continuously done in less amount and there is always a synthesis of less quantity of protein.
The Lac Operon consist of three genes viz. lac Z, lac Y, and lac A that participate in the catabolism of the disaccharide lactose.
- lac Z is β – galactosidase, an enzyme that cleaves lactose in galactose and glucose.
- lac Y is permease which transports lactose from the extracellular environment into the cell. Along with lactose, TONP- G ( toxic galactoside) comes into the cell through lactose permease.TOP- G is inactivated by thiogalactoside transacetylase by adding the Acetyl group.
- lac A is a thiogalactosidase transacetylase that transfers acetyl groups from acetyl -CoA to lactose.
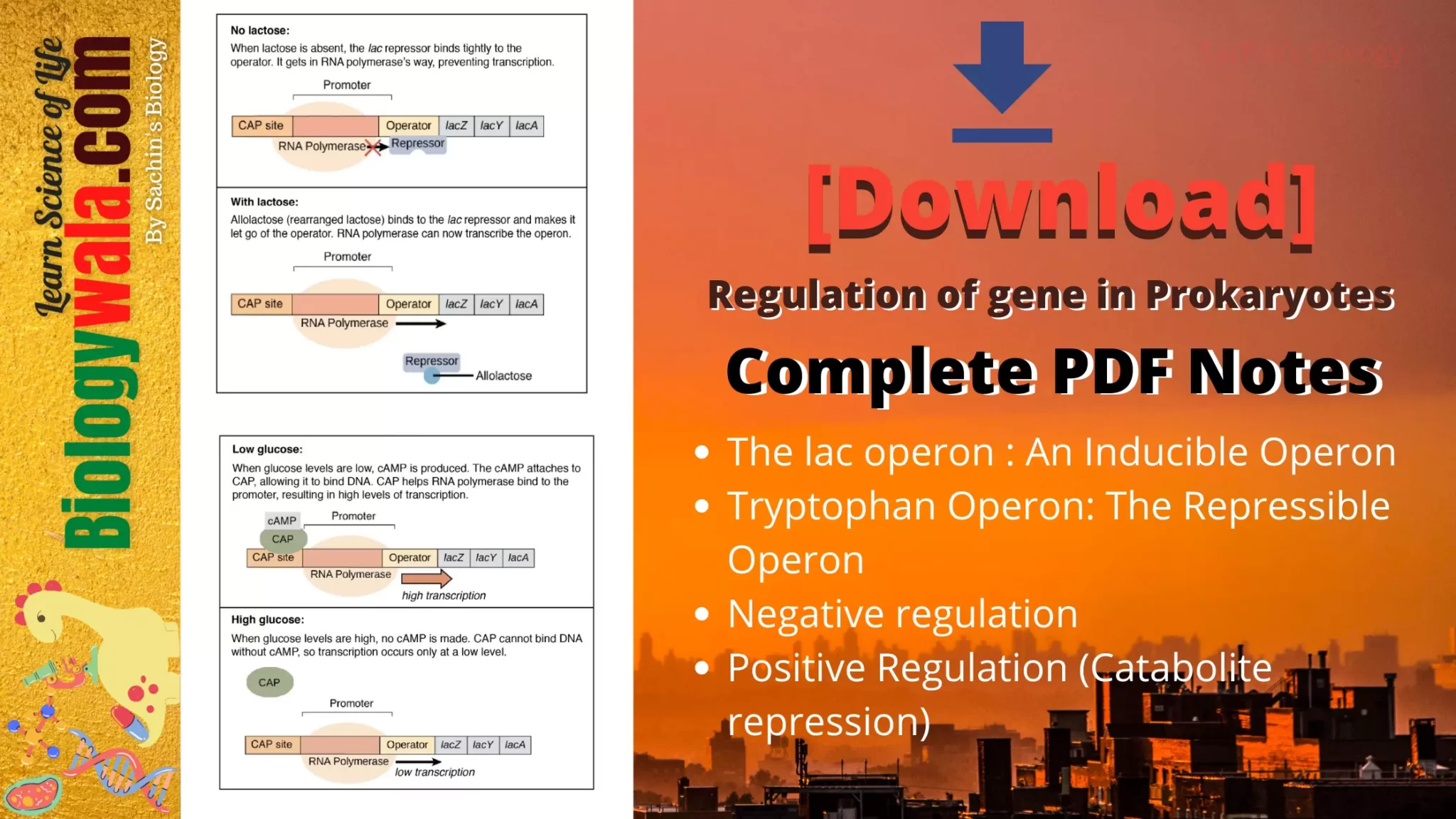
- The promoter is the enzyme which controls the expression of three structural genes (i.e. lac Z, lac Y, lac A). These three different structural genes’ expression is controlled by a single promoter which is called as “Polycistronic “ expression of the gene.
- The operator acts as a binding site for a repressor protein that blocks RNA polymerase.
- Catabolite Activator Protein (CAP ) is the transcriptional activator which binds to cAMP to activate the transcription of lac Operon by RNA polymerase. When glucose level decreases, cyclic AMP (cAMP ) begins to accumulate in the cell.
- lac I am the regulatory gene of the lac operon called a lac repressor. It can prevent transcription and can recognize and bind to the inducer.
Lactose is the inducer that binds to the repressor to form a repressor-inducer complex and prevents the repressor from binding to the operator.
Negative regulation :
The Repressor lac I gene is a homotetramer having four identical subunits that make the DNA binding domain and the other two subunits make the Inducer binding domain.
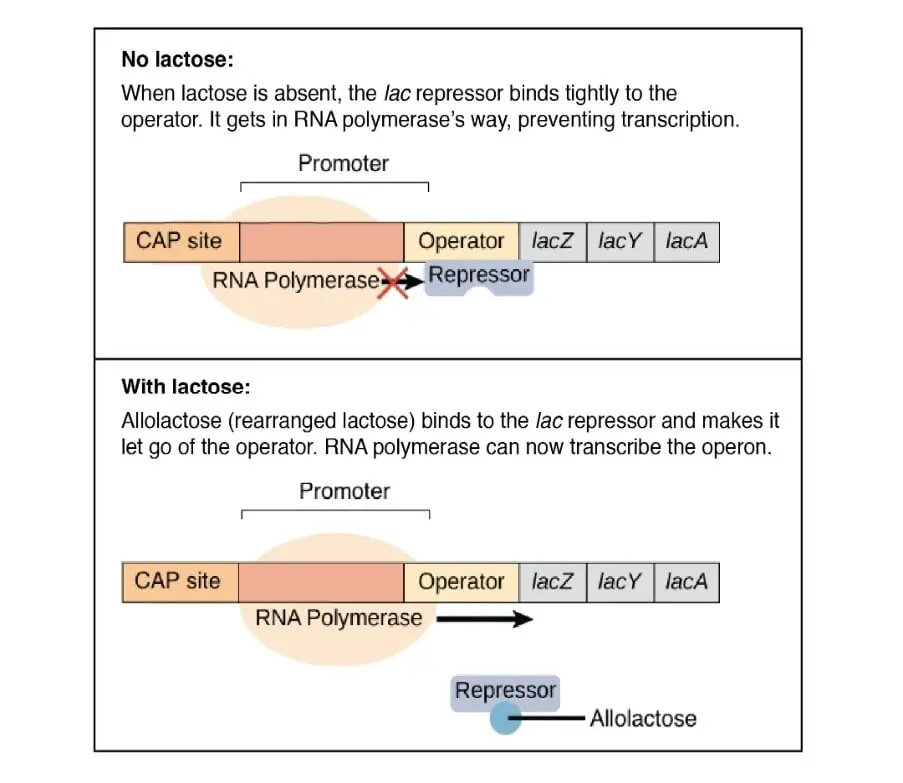
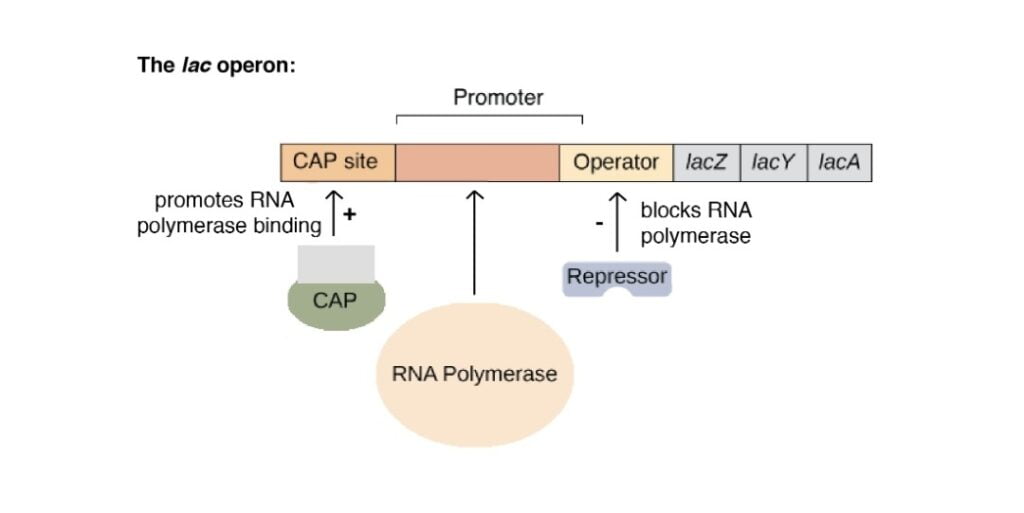
lactose is absent lac repressor (lac I) is active and binds to the Operator and interferes with the initiation of transcription. So in this case transcription rate (TR) is less than the basal transcription rate (BTR).(TR < BTR)
- If lactose is present it gets isomerize into allolactose .The. Lac repressor has high affinity for allolactose.This allolactose gets bind to the repressor and form inducer – repressor complex , preventing repressor from binding to the Operator region and thus allowing RNA polymerase to transcribe the genes .In this case , Transcription rate is equal to the basal transcription rate (BTR) i.e.TR = BTR .
Positive Regulation (Catabolite repression):
- Glucose is the preferred source of energy for E.coli.Bacteria will Consume available glucose before any other carbon source.
- Glucose leads to repression of expression of lactose and this is called as “Catabolite Repression”.
- If glucose is present in cell the cAMP inside the cell decreases due to inactive adenylate cyclase .( Enzyme adenylate cyclase synthesize the cAMP).
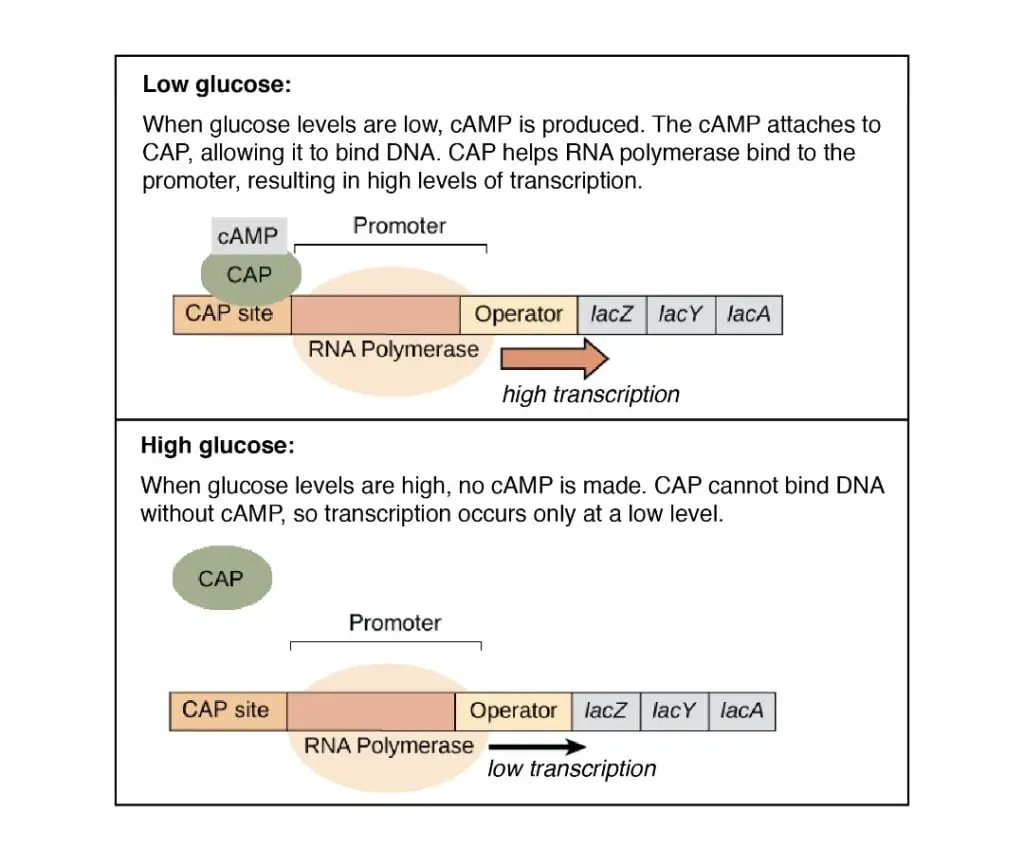
However, when there is little glucose in the cell, the level of cAMP is increased and binds to the active CAP forming cAMP -CAP complex which then binds to DNA at specific sites. This Binding of cAMP – CAP to the DNA site will enhance the efficiency of transcription.In this case, transcription rate (TR) increases than basal transcription rate (BTR) i.e.TR > BTR.
{ Glucose↑ – cAMP ↓ – CAP Inactive
Glucose ↓ – cAMP ↑ – CAP active – TR increases}
- Glucose absent & lactose absent :
Due to the absence of lactose, there is no transcription of lac Operon. Glucose levels are low so cAMP levels are increased, so CAP is active and bound to DNA. As lactose is absent means there is negative regulation so the transcription rate is very less than the basal transcription rate (TR <<<<BTR).
- Glucose present & lactose absent :
No transcription of lac Operon.cAMP levels are also low due to high levels of glucose, so CAP is inactive and cannot bind DNA due to the absence of lactose, lac operon remains off and transcription rate is less than basal transcription rate (TR<<<<BTR ).
- Glucose absent & lactose present:
Lactose is present, so strong transcription of the lac operon occurs .cAMP levels are high due to the absence of glucose, so lac operon is positively regulated. In this case, the Transcription rate is more than the basal transcription rate.(TR>>>>BR).
- Glucose present & lactose present:
The lac repressor is released from the operator and bind to the Inducer (allolactose). However, cAMP levels are low due to the presence of glucose. Thus CAP remains inactive and cannot bind to DNA.
The transcription rate is equal to the basal transcription rate because transcription only occurs at a low or leaky level (TR = BTR).
| Glucose | Lactose | Level of transcription | Rate of transcription | |
| Case I | – | – | No transcription | TR<<<<BTR |
| Case II | + | – | No transcription | TR<<<<BTR |
| Case III | – | + | Strong transcription | TR>>>>BTR |
| Case IV | + | + | Low-level transcription | TR = BTR |
Confirmatory expression by Jacob and Monad:-
- IPTG induction is a method of regulating protein synthesis by activating transcription of the lac operon. IPTG (Isopropyl β – D -1-thiogalactopyranoside) is a gratuitous inducer which is an analog of allolactose which bind to the repressor lac I .
- Lactose can be degraded in cell, while IPTG can not and induces the lac – operon expression.When IPTG induced in cell ,it binds to the repressor and because of its non – degrading properties there is continuous synthesis of protein. However, when lactose is bind to repressor it will get degrade after some time and repressor will bind to operator blocking the activity of RNA polymerase.
- In Blue- white screening method,IPTG is used along with X- gal(5 – bromo- 4 -chloro-3-indolyl -β-D-galactopyranoside) ,IPTG induces the expression of lac Z gene,it’s not a substrate for β-galactosidease but an Inducer . X- gal us required for visual indication of expression of a functional β- galactosidase enzyme.
- X-gal is hydrolyzed by enzyme β-galactosidase breaking the β- glycosidic bond in D- lactose and it gives blue – colour.This confirms that lac operon works in it.
Mutation in lac operon –
- In non – mutations state,if lac Z is a normal gene it is written as lac Z+ similarly with lac Y+ and lac A+ .’+’ means all are “wild” type and doing perfectly normal function.
- In mutation state lac ‘Z-’means the gene is non – functional and unable to produce the enzyme similarly with lac ‘Y-’ and lac ‘A-’ gene.
- ‘I-’means lac repressor gene has some mutation as a result of it there no lac repressor.If there is no lac repressor it doesn’t matter there is presence or absence of lactose,the lac operon will always turn on.
- Constitutive mutation– In constitutive mutation gene product is continuously produced and there is no control over it’s expression.
- Cis- acting genes – Promoter gene (P) and operator gene(O)are the part of the regulatory gene and not producing any product so they are unable to move from their respective Location.
- Trans- acting genes- lac I ,lac Z ,lac Y and lac A are trans acting genes.As all genes produce a particular product.So they can move from one place to other .
- Oc is the cis- mutation in which operator change its structure in such a way that the lac repressor fail to bind it ,so the lac operon will always be turned on .
- Super repressor (Is):– The inhibitor of repressor (allolactose) can not bind to the repressor due to structural changes in the binding site of lactose,so lactose can not inhibit the repressor as a result the repressor is always active and the lac Operon will always be turned off.
- Mero- diploids:- When a cell carrying a second copy of some or all the lac operon genes these cells are called Mero- diploids .E.coli become diploid for lac operon.Only lac Operon is diploid hence it is called as partly diploid i.e.mero- diploid .
In mero – diploids it is not important who secrete repressor or expressed β- galactosidase if one of them can express gives result positive or negative.
I+ P+ O+ Z- Y+ A+
In absence of lactose, lac operon will be off and there is no expression of genes.
But in the presence of lactose, lac operon will be turned on but lac Z is non-functional so there will be no expression of the gene.
Is O+ P+ Z+
F’ I- Oc P+ Z+
Both in the presence or absence of lactose, there will be a constitutive expression as Oc is a constitutive mutant. So there is no effect of Is.
lac I+ O+ P+ Z+
F’ lac I- O+ P+ Z-
In absence of lactose, lac I acts as a transactive factor and inhibit expression on both E.coli lac operon and F’ lac operon. In presence of lactose, the repressor will bind to lactose and the lac operon will activate.
Regulation of gene in Prokaryotes: Tryptophan Operon
The Repressible Operon
- It is a type of prokaryotic gene regulation process which is related with biosynthesis of tryptophan amino acid .
- This is an anabolic operon pathway. In Trp operon after the promoter and operator, there is a stretch of DNA sequence known as leader sequence (101nt). Right after that there are five structural elements E, D, C, B, and A which codes for five different types of protein.
- Trp E – Anthranilate synthase
2. Trp D – Anthranilate transferase
3. Trp C – Anthranilate isomerase
4. Trp B – Tryptophan synthase -β
5. Trp A – Tryptophan synthase- α
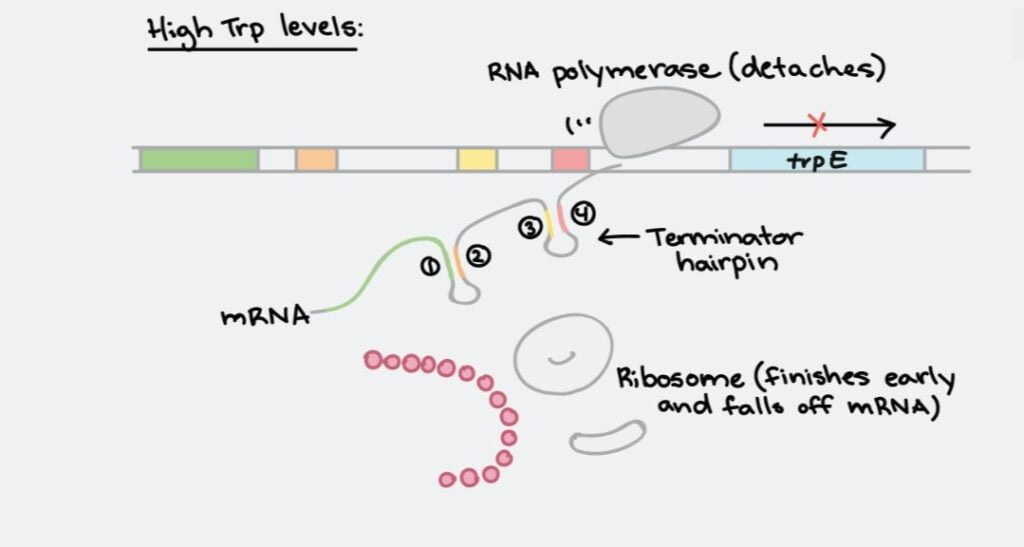
- It also contains a repressive regulator gene called trpR. The demand for Tryptophan in the cytoplasm is very less .hence this Operon is mostly present in a repressed form called a repressible operon.
- Trp codes for repressor but alone repressor protein can’t bind to the operator site. When trp binds to the repressor It binds to the operator site and the Operon gets repressed.
- Generally, normal operon repressors repress 100 times more than the normal transcription rate. But in Tryptophan Operon repressor carries out 1000 times repression. So excess repression takes place in Trp operon.
- This is done by repression of the basal level of transcription to avoid loss of energy.
- In absence of Tryptophan,trpR gets inactive and can’t bind to the operator so the operon will turn on and starts the synthesis of Tryptophan.
- In presence of Tryptophan, the inactive repressor protein called aporepressor needs corepressor to get active. This active complex get binds to the operator which blocks the transcription.
- In the Attenuation process, rather than blocking the initiation of transcription, attenuation averts the completion of transcription.
- When Tryptophan is high, the ribosome moves quickly with the leader sequence and forms a termination loop or hairpin loop and Transcription ends.
- When Tryptophan levels are low, the ribosome moves slowly along with the leader sequence forming the antiterminator hairpin loop. This allows the transcription of trp operon.
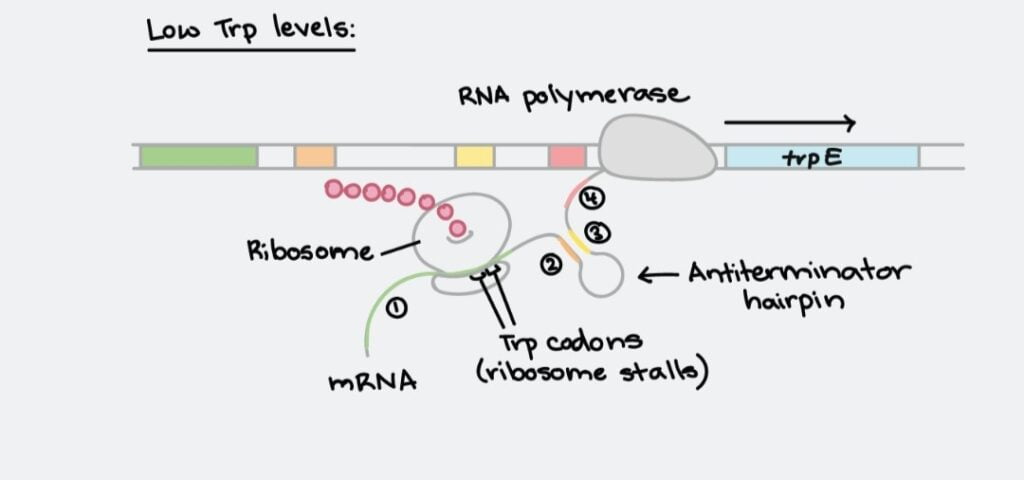
- In other words, high levels of Tryptophan in cells turn off the expression of the operon. Since the cell does not need more synthesis of Tryptophan.
This is it for Download Regulation of gene in Prokaryotes complete pdf notes article today hope you like it. If you really believe there is something missing in this article then please let us know in the comment box below or tell us on Instagram.
Also like:
[DOWNLOAD] Bruce Albert’s Molecular Biology of The Cell book PDF
[Download] The Biology of Cancer Weinberg PDF Book 2nd edition
[Download] Top 5 Books for Molecular Biology: Basic to Advance
Download Free Reference Books for Genetics
If you want important notes and updates about exams on your mobile then you can join SACHIN’S BIOLOGY on Instagram or Facebook and can directly talk to the founder of Sachin’s Biology and Author of biologywala.com Mr.Sachin Chavan M.Sc. NET JRF (AIR 21) GATE!


4 Responses
[…] control of gene expression in eukaryotes is more complicated than the gene expression in prokaryotes. The pathway for a gene to protein is much more complex in eukaryotes than the pathway in […]
[…] Regulation of gene in Prokaryotes […]
[…] control of gene expression in eukaryotes is completely different from the gene expression in prokaryotes. The pathways that produce gene is more complex in eucaryotes than the pathway in prokaryotes. This […]
[…] Regulation of gene in Prokaryotes […]